FACES OF A*STAR
Epstein-Barr virus rewires host epigenomes to drive stomach cancer
- Researchers in Japan and Singapore have discovered a molecular mechanism that explains how Epstein-Barr virus (EBV) infection alters a host’s epigenome to promote tumorigenesis (the transformation of normal cells into cancer cells) in certain types of stomach cancer.
- The findings suggest that EBV infection plays an important role in the development of EBV-associated stomach cancers, and provide fresh insights that may lead to new therapies for stomach and other virus-related malignancies.
SINGAPORE, 27 July 2020 – The Epstein-Barr virus (EBV), one of the most common human viruses, is associated with about 8–10 per cent of stomach — or gastric — cancers, the third leading cause of cancer death globally. Researchers from Chiba University in Japan, Duke-NUS Medical School, Singapore, and the Agency for Science, Technology and Research (A*STAR)’s Genome Institute of Singapore (GIS) have revealed a novel paradigm in EBV-associated gastric cancer, whereby the EBV viral genome directly alters the host epigenetic landscape to promote the activation of proto-oncogenes (genes involved in normal cell growth that can mutate into cancer-causing genes) and tumorigenesis.
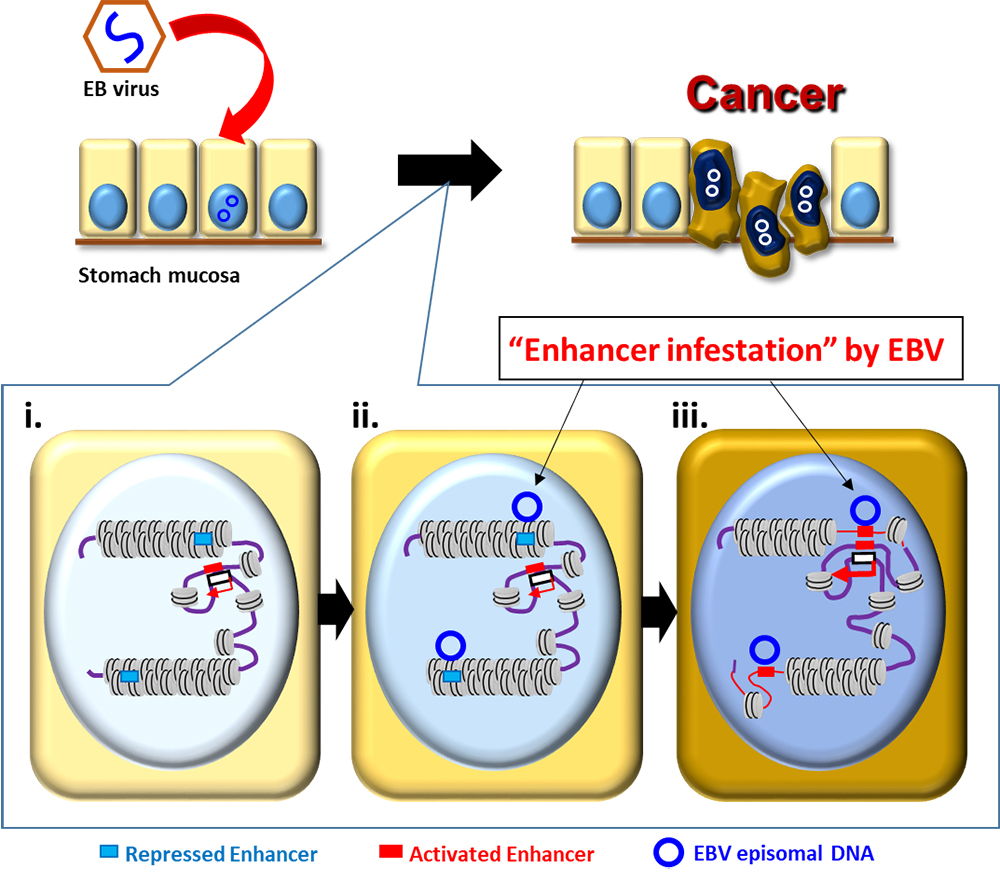
Credit: Chiba University | Duke-NUS Medical School | Genome Institute of Singapore
Schematic model of Epstein-Barr virus (EBV) enhancer infestation driving heterochromatin, epigenomic and transcriptional rewiring in EBV-positive gastric cancers.
- Enhancers silenced in the closed heterochromatin region (pale blue box).
- EBV DNA bound to the closed regions (blue circle).
- Rewiring from inactive to active state where the closed regions are opened, the silenced enhancers are activated, and the neighbouring proto-oncogenes are activated.
The human genome is the complete set of human genetic information, and the epigenome describes modifications to the genome that determine whether genes are turned on or off when and where they are needed. Unlike genetic information, the epigenome is dynamic and responsive to external stimuli; certain external stimuli can cause abnormal DNA modifications which, in turn, can disrupt normal gene expression and contribute to cancer development.
The research group, led by senior and co-corresponding authors, Dr Atsushi Kaneda, Professor at the Graduate School of Medicine, Chiba University, and Dr Patrick Tan, Professor at the Programme in Cancer and Stem Cell Biology, Duke-NUS Medical School, and Executive Director of GIS, conducted a comprehensive analysis of three-dimensional genomic structures in human cells. These ranged from gastric cancer cell lines, patient samples, normal gastric epithelial cells, and EBV-associated gastric cancer. Combined with virus infection analyses, the researchers found abnormally activated genomic regions specific to EBV-positive stomach cancer. Experimental EBV infection of cultured stomach cells reproduced the phenomena of EBV binding to these inactive and closed genomic regions and their abnormal activation.
‟Cells put active marks on genomic regions necessary for their behaviours and utilise them, and inactive marks on unnecessary genomic regions that are tightly closed and not to be utilised,” explained Prof Kaneda. “We made the striking observation that strong inactive marks were lost in specific genomic regions when we infected stomach cells with EBV.”
The researchers further found that genetic enhancers (short pieces of DNA that help encourage genes to make proteins) ‘silenced’ in the closed regions were activated by the virus to upregulate nearby cancer-related genes, leading to the proliferation of cancerous cells. This ‘enhancer infestation’ model, as the researchers termed it, reveals a novel mechanism of tumorigenesis that does not require genetic alterations, and instead works by reprograming the epigenetic landscape of human cells to convert latent enhancers from a silenced to an active state.
Prof Patrick Tan, who is also a member of the Singapore Gastric Cancer Consortium, remarked, ‟In all EBV-positive stomach cancer cells and primary stomach cancer patient samples studied, EBV DNA bound to largely the same genomic regions that also showed abnormal activation. These same regions also changed from inactive to active states by experimental EBV infection.”
This mechanism of ‘enhancer infestation’ led to the activation of neighbouring proto-oncogenes in human cells and it is likely to contribute to EBV-associated oncogenesis in multiple cancer cell types. Notably, the researchers also found that, even after eliminating EBV genomes, epigenetic modifications that were induced continued to persist, suggesting a ‘hit-and-run’ mechanism in which, once an EBV episome alters the chromatin topology of human cells, these altered topologies are stable and persist even after removal of the EBV episome.
Prof Kaneda reiterated, “While 8–10 per cent of stomach cancer is associated with EBV, we believe our enhancer infestation model provides a new mechanism of cancer involving epigenomic alterations and viral infection that may be relevant to a broader range of cancers and associated diseases.”
Prof Tan added, “Infections by EBV are estimated to cause over 200,000 cancers per year worldwide, including certain stomach cancers. Our study highlights new potential drug targets in EBV-positive malignancies, revealed by epigenetics and previously invisible using more conventional genetic sequencing studies.”
###
Reference: Okabe, A., Huang, K.K., Matsusaka, K. et al. Cross species chromatin interactions drive transcriptional rewiring in Epstein Barr virus positive gastric adenocarcinoma. Nature Genetics (2020). https://doi.org./10.1038/s41588-020-0665-7 (Direct link to the paper)
About Duke-NUS Medical School
Duke-NUS is Singapore’s flagship graduate entry medical school, established in 2005 with a strategic, government-led partnership between two world-class institutions: Duke University School of Medicine and the National University of Singapore (NUS). Through an innovative curriculum, students at Duke-NUS are nurtured to become multi-faceted ‘Clinicians Plus’ poised to steer the healthcare and biomedical ecosystem in Singapore and beyond. A leader in ground-breaking research and translational innovation, Duke-NUS has gained international renown through its five signature research programmes and eight centres. The enduring impact of its discoveries is amplified by its successful Academic Medicine partnership with Singapore Health Services (SingHealth), Singapore’s largest healthcare group. This strategic alliance has spawned 15 Academic Clinical Programmes, which harness multi-disciplinary research and education to transform medicine and improve lives.
For more information, please visit www.duke-nus.edu.sg
About Chiba University Graduate School of Medicine
Chiba University was founded in 1949, unifying several regional former national colleges and schools. Since then, pursuing the goals of excellence has resulted in Chiba University becoming one of the leading academic research centres of Japan.
The history of Chiba University Graduate School of Medicine can be traced back as early as 1874, when its predecessor was founded. Chiba University Graduate School of Medicine focuses on the promotion of research in "Innovative Therapeutics", a new field that connects basic and clinical medicine.
Chiba University offers “Innovative Medicine CHIBA Doctoral WISE Program (iMeC-WISE)” supported by the Japanese government. Based on the 100-year history of the fields of medicine and pharmaceutical sciences, iMeC-WISE aims to foster the next generation of world-class researchers and innovators, who will contribute to the development of medical sciences, pave the way to novel therapies and drugs, and develop sustainable healthcare systems.
For more information about Chiba University
About A*STAR’s Genome Institute of Singapore (GIS)
The Genome Institute of Singapore (GIS) is an institute of the Agency for Science, Technology and Research (A*STAR). It has a global vision that seeks to use genomic sciences to achieve extraordinary improvements in human health and public prosperity. Established in 2000 as a centre for genomic discovery, the GIS pursues the integration of technology, genetics and biology towards academic, economic and societal impact, with a mission to “read, reveal and write DNA for a better Singapore and world”.
Key research areas at the GIS include Precision Medicine & Population Genomics, Genome Informatics, Spatial & Single Cell Systems, Epigenetic & Epitranscriptomic Regulation, Genome Architecture & Design, and Sequencing Platforms. The genomics infrastructure at the GIS is also utilised to train new scientific talent, to function as a bridge for academic and industrial research, and to explore scientific questions of high impact.
For more information about GIS, please visit www.a-star.edu.sg/gis.
About the Agency for Science, Technology and Research (A*STAR)
The Agency for Science, Technology and Research (A*STAR) is Singapore's lead public sector R&D agency, spearheading economic-oriented research to advance scientific discovery and develop innovative technology. Through open innovation, we collaborate with our partners in both the public and private sectors to benefit society.
As a Science and Technology Organisation, A*STAR bridges the gap between academia and industry. Our research creates economic growth and jobs for Singapore, and enhances lives by contributing to societal benefits such as improving outcomes in healthcare, urban living, and sustainability.
We play a key role in nurturing and developing a diversity of talent and leaders in our Agency and research entities, the wider research community and industry. A*STAR’s R&D activities span biomedical sciences and physical sciences and engineering, with research entities primarily located in Biopolis and Fusionopolis. For ongoing news, visit www.a-star.edu.sg.
Was this article helpful?
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
