Innovative Metagenomics Study Maps Out Antibiotic Resistance Genes From Hospital Environment Microbiomes
Findings were made by A*STAR’s Genome Institute of Singapore (GIS), through its analysis of genetic sequences of microbes and multi-drug resistant organisms found in hospital environments
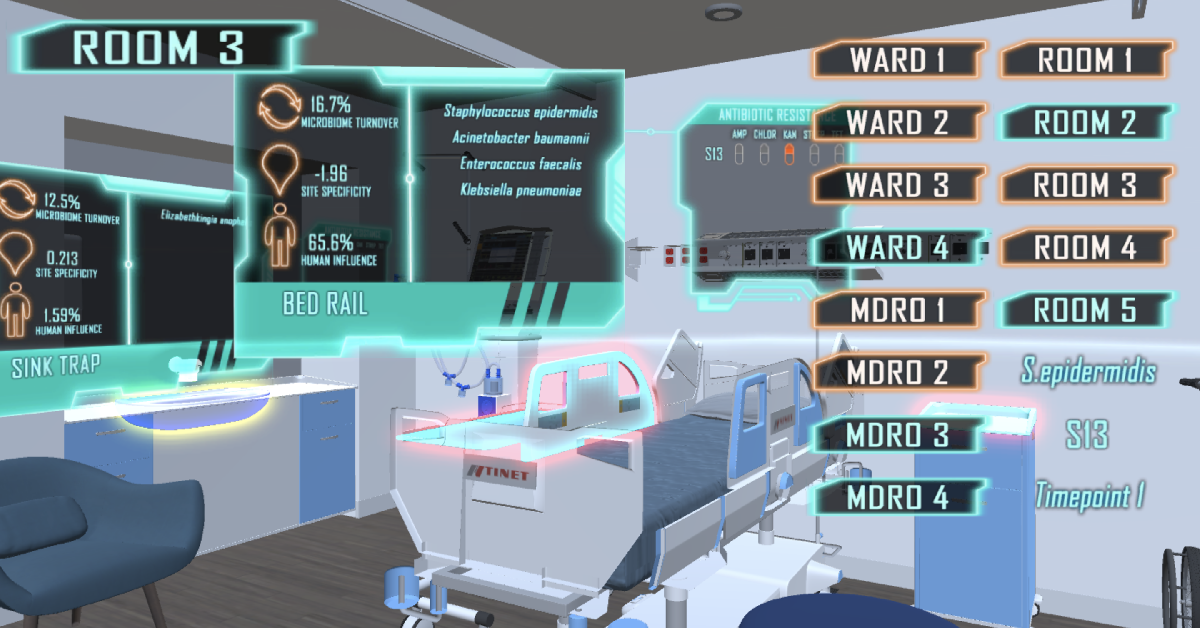
Virtual-reality visualisation of metagenomic data for infection control in smart hospitals (Copyright: A*STAR’s Genome Institute of Singapore)
Singapore – Nature Medicine published a study by the Agency for Science, Technology and Research’s (A*STAR) Genome Institute of Singapore (GIS), on 8 June 2020, providing the first extensive genomic mapping of microbiomes and antibiotic resistance genes in a tertiary hospital in Singapore.
This paper is a potentially ground-breaking analysis, enlisting the emerging technology of metagenomics to provide a detailed survey useful in the increasingly important worldwide battle against antimicrobial resistance, and represents a collaboration between GIS, Tan Tock Seng Hospital (TTSH), National Centre for Infectious Diseases (NCID), National University Hospital, Singapore General Hospital (SGH), Weill Cornell Medicine, and the MetaSUB Consortium1.
This study maps out for the first time the diversity and range of bacteria and antibiotic resistant genes found in hospitals. In this study, environmental samples were collected from different sites in a hospital, cultures were enriched to find antibiotic resistant bacteria, and the sequences were compared to databases of previously published sequences from bacteria found in hospital environments as well as patients.
While all-out efforts have been focused on the COVID-19 pandemic, the global epidemic of antibiotic resistance is a looming danger, projected to cause millions of deaths worldwide over the next three decades, with an economic impact estimated at 100 trillion dollars by 20502. Antimicrobial resistance will exact a heavy healthcare burden in both developed and developing countries3.
This study shows that modern gene sequencing technologies and metagenomic analyses can systematically characterise the distribution of bacteria, along with antibiotic resistance genes. Such detailed data could provide potential information to identify microbial reservoirs in hospital environments, including organisms that form biofilms or are potential risks for human infection.
Dr Niranjan Nagarajan, Associate Director and Group Leader at GIS, said, “Our analysis highlights that hospital environments may harbour significant uncharacterised genetic diversity. A large baseline survey such as this study provides a reference map that can be updated based on periodic scans.”
Associate Prof Christopher E Mason, from Weill Cornell Medicine, commented, “We are now merging these data with the global catalogue of other viruses, antibiotic markers, fungi, and bacteria found in the global MetaSUB Consortium to give us a genetic ranking for antibiotic resistance that we can use for tracing in hospitals around the world.”
“This study leverages cutting-edge metagenomics to understand the distribution of multidrug-resistant organisms. The current findings support the possibility of precision infection control where we can apply targeted infection control interventions to control MDROs in the healthcare setting,” said Dr Kalisvar Marimuthu and Associate Professor Ng Oon Tek, Senior Consultants at NCID.
Prof Patrick Tan, Executive Director of GIS, said, “These findings highlight the importance of characterising antibiotic resistance reservoirs in the hospital environment. They establish the feasibility of systematic genomic surveys to help target resources more efficiently for preventing hospital-acquired infections.”
Ongoing work by the team will explore how RNA viruses might be distributed and persist in the hospital environment, the impact of various cleaning measures on their distribution, and how this may improve care for hospitalised patients.
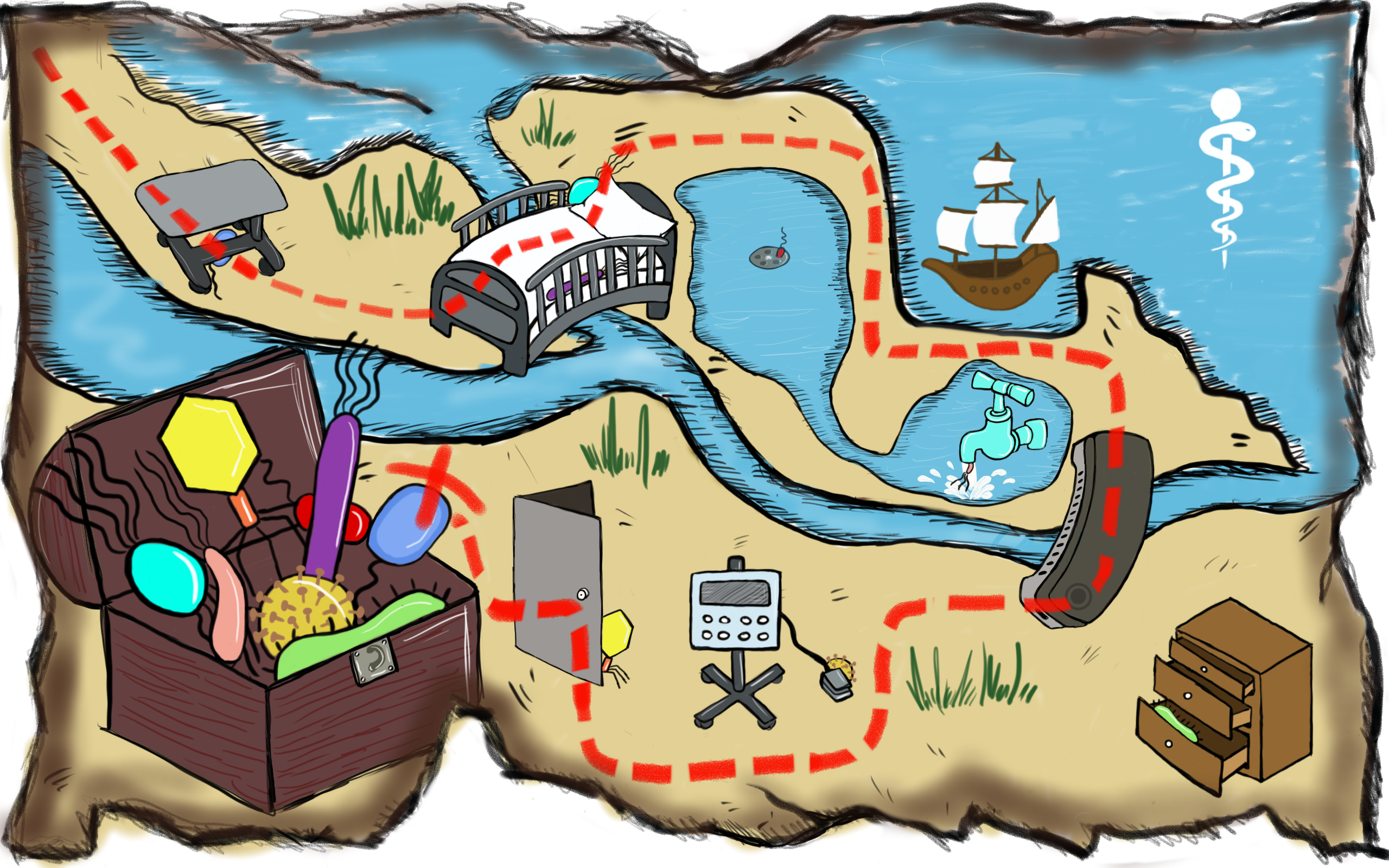
Charting out a map of opportunistic pathogens in the hospital environment (Copyright: A*STAR’s Genome Institute of Singapore)
– END –
1 MetaSUB Consortium: The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International consortium is a novel, interdisciplinary initiative made up of experts across many fields, including genomics, data analysis, engineering, public health, and design.
2 O’Neill, J. Tackling drug-resistant infections globally: final report and recommendations, <https://amr-review.org/Publications.html>
Enclosed:
About A*STAR's Genome Institute of Singapore (GIS)
The Genome Institute of Singapore (GIS) is an institute of the Agency for Science, Technology and Research (A*STAR). It has a global vision that seeks to use genomic sciences to achieve extraordinary improvements in human health and public prosperity. Established in 2000 as a centre for genomic discovery, the GIS will pursue the integration of technology, genetics and biology towards academic, economic and societal impact.
The key research areas at the GIS include Human Genetics, Infectious Diseases, Cancer Therapeutics and Stratified Oncology, Stem Cell and Regenerative Biology, Cancer Stem Cell Biology, Computational and Systems Biology, and Translational Research.
The genomics infrastructure at the GIS is utilised to train new scientific talent, to function as a bridge for academic and industrial research, and to explore scientific questions of high impact.
For more information about GIS, please visit https://www.a-star.edu.sg/gis.
About the Agency for Science, Technology and Research (A*STAR)
The Agency for Science, Technology and Research (A*STAR) is Singapore's lead public sector agency that spearheads economic oriented research to advance scientific discovery and develop innovative technology. Through open innovation- we collaborate with our partners in both the public and private sectors to benefit society.
As a Science and Technology Organisation- A*STAR bridges the gap between academia and industry. Our research creates economic growth and jobs for Singapore- and enhances lives by contributing to societal benefits such as improving outcomes in healthcare- urban living- and sustainability.
We play a key role in nurturing and developing a diversity of talent and leaders in our Agency and Research Institutes- the wider research community and industry. A*STAR oversees 18 biomedical sciences and physical sciences and engineering research entities primarily located in Biopolis and Fusionopolis. For ongoing news, visit www.a-star.edu.sg.
ANNEX A – NOTES TO EDITOR
The research findings described in this media release can be found in the scientific journal Nature Medicine, under the title, “Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment” by Kern Rei Chng1,*, Chenhao Li1,*, Denis Bertrand1,*, Amanda Hui Qi Ng1, Junmei Samantha Kwah1, Hwee Meng Low1, Chengxuan Tong1, Maanasa Natrajan1, Michael Hongjie Zhang 1, Licheng Xu2, Karrie Kwan Ki Ko3,4,5, Eliza Xin Pei Ho1, Tamar V. Av-Shalom1, Jeanette Woon Pei Teo6, Chiea Chuen Khor1, MetaSUB Consortium, Swaine L. Chen1 , Christopher E. Mason7, Oon Tek N8, Kalisvar Marimuthu8, Brenda Ang8, Niranjan Nagarajan1,9,#.
- Genome Institute of Singapore, 60 Biopolis Street, #02-01 Genome, Singapore 138672, Singapore
- Singapore University of Technology and Design, 8 Somapah Rd, Singapore 487372, Singapore
- Department of Microbiology, Singapore General Hospital, Singapore 169856, Singapore
- Department of Molecular Pathology, Singapore General Hospital, Singapore 169856, Singapore
- Duke-NUS Graduate Medical School, Singapore 169857, Singapore
- Department of Laboratory Medicine, National University Hospital, 5 Lower Kent Ridge Road, Main Building I, Singapore 119074, Singapore
- Department of Physiology and Biophysics, Weill Cornell Medicine, New York 10065, USA
- Institute of Infectious Diseases and Epidemiology^, Tan Tock Seng Hospital, 11 Jalan Tan Tock Seng, Singapore 304833, Singapore
- National University of Singapore, 21 Lower Kent Ridge Road, Singapore 119077, Singapore
* Contributed equally
# Corresponding author
^ Now known as the National Centre for Infectious Diseases
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
