Fragle: Affordable AI-Powered Cancer Monitoring

Detecting cancer relapse months earlier and at a fraction of the cost
Current cancer monitoring methods are costly and often detect relapse only after tumours have become visible. Tests may cost thousands of dollars per run, making them impractical for frequent use. At A*STAR, researchers studied DNA fragment-length patterns in plasma — a field known as fragmentomics — and discovered they reveal tumour activity even at very low levels. From this research, they developed Fragle, an AI-powered tool that quantifies circulating tumour DNA (ctDNA) rapidly and at a fraction of the cost.
Fragle works with ultra-low-pass whole-genome sequencing or targeted panels already in hospitals, returning results in under a minute. Validated in a study featured in Nature Biomedical Engineering (March 2025), it can measure ctDNA fractions as low as 1%. The National Cancer Centre Singapore is now applying it in an ongoing trial with more than 100 lung cancer patients, monitored every two months to track ctDNA dynamics and detect relapse earlier than standard imaging. By lowering costs and simplifying workflows, Fragle makes ctDNA analysis practical for routine use, enabling earlier intervention and more equitable access to personalised oncology.
For enquiries and collaboration opportunities, please use our contact form.
Holistic Wound Care: Multiplexed Sensor × Acelevaé™ Hydrogel
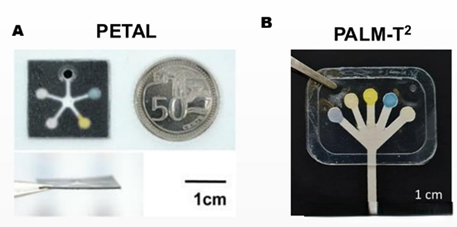

Smarter monitoring and advanced treatment for chronic wound management
Chronic wounds such as diabetic foot ulcers affect millions worldwide and cost healthcare systems billions each year. Healing is often hindered by slow diagnosis and poor tissue repair. Two Singapore-developed innovations are addressing these challenges with smarter diagnostics and more effective therapies.
The Multiplexed Wound Sensor Patch, developed at A*STAR in partnership with the National University of Singapore (NUS), is a low-cost disposable patch that measures multiple infection and inflammation biomarkers directly at the wound site. Using a colour-based sensing design with mobile imaging and NUS algorithms, it delivers a bedside readout in under 15 minutes. A clinical study is underway at Sengkang General Hospital.
The Product Ten (PTEN) Hydrogel, co-developed by A*STAR and Nanyang Technological University (NTU), is an antioxidant-rich formulation that reduces inflammation and supports tissue repair. Previously known as Product Ten Hydrogel, the technology has now been spun off as Acelevaé™ under an A*STAR spin-off Argonaute Lifesciences and is undergoing evaluation at Changi General Hospital. Together, these technologies form a comprehensive solution that could transform wound care and expand access to effective treatment.
For enquiries and collaboration opportunities, please use our contact form.
Restoring the Skin Barrier: A Novel Biogenic Amine Technology
Targeting the root causes of skin fragility
As skin ages, it becomes thinner, weaker, and slower to repair. This fragility increases health risks for the elderly and worsens quality of life. Existing creams only lock in moisture temporarily, without addressing underlying biological causes.
Researchers at A*STAR and the National Skin Centre (NSC) identified that an imbalance of biogenic amines which are natural molecules essential for skin function, play a key role in barrier dysfunction and poor repair. Building on this insight, they developed a plant-based formulation that restores biogenic amine balance, strengthening the barrier from within.
Laboratory tests on advanced skin models showed improved barrier thickness, enhanced tissue repair, and reduced pigmentation. Two topical cream prototypes have already been formulated and are moving toward clinical evaluation. By targeting the root causes of skin fragility, this innovation offers a regenerative approach that goes beyond temporary moisturisation. It has the potential to provide more durable protection and repair for ageing skin.
For enquiries and collaboration opportunities, please use our contact form.
SNPdrug3D: Mapping Genetic Variants to Drug Response in 3D
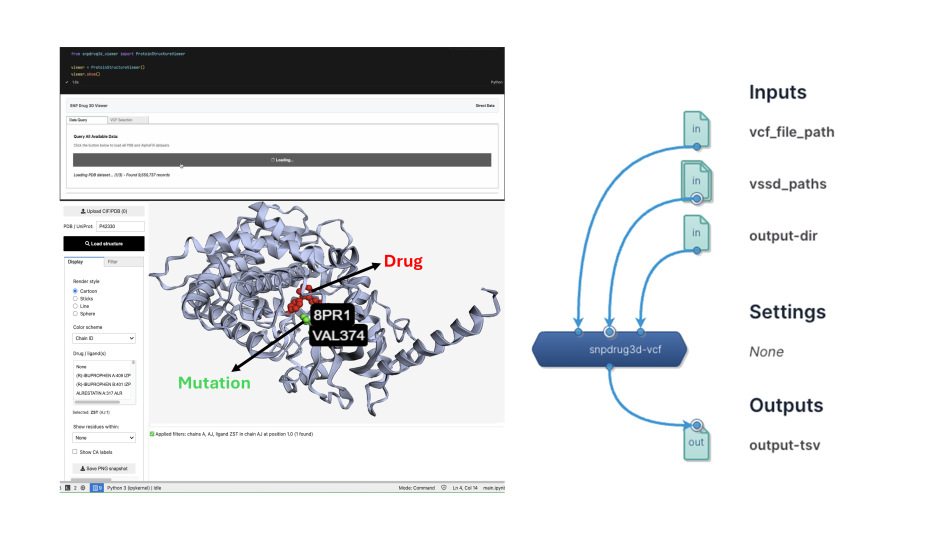
Turning genetic data into actionable insights for patient care
Researchers at A*STAR have developed deep expertise in structural bioinformatics, harnessing protein structures to reveal how molecules interact. Building on this foundation, and in partnership with Temus under the joint A*STAR–Temus Lab initiative, the team created SNPdrug3D, an AI-powered platform designed to tackle a key challenge in precision medicine: interpreting how genetic variants influence how patients respond to treatment.
SNPdrug3D uses three-dimensional structural models to show how these variants affect drug binding. It is the largest platform of its kind, covering more than 70 million protein-altering variants, 200,000 protein structures (experimental and AI-predicted), and nearly 1,800 drugs. This has generated over 4 million variant–drug mappings, enabling clinicians and researchers to uncover relationships that were previously hidden.
Already in clinical use in Singapore, oncologists and specialists are applying SNPdrug3D to interpret sequencing data for cancer, eye, and autoimmune patients. It also incorporates Singapore’s population-level genomic data to support patient stratification and national precision medicine efforts. Beyond individual cases, SNPdrug3D can perform pharmacogenetic assessments of personal genomes, pinpointing variants that may disrupt binding and influence treatment.
Deployed on Illumina’s commercial cloud and compatible across platforms, including as a Python package for researchers, SNPdrug3D shows how deep R&D at A*STAR has been translated into a scalable tool that advances personalised medicine both locally and globally.
For enquiries and collaboration opportunities, please use our contact form.




