A*STAR NEWS
PRESS RELEASES
Fishing for Cellular Identities
Scientists develop ground-breaking split-FISH technology that is able to generate highly detailed molecular maps of cells and tissues
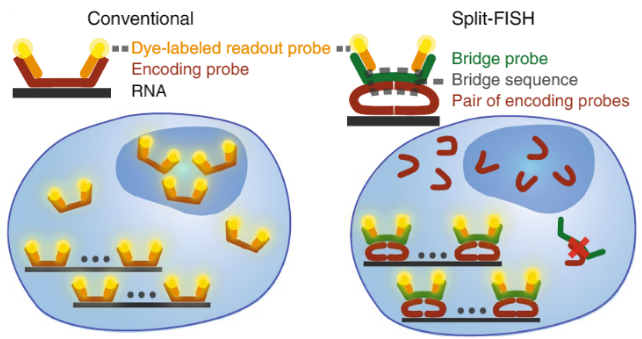
Schematic comparison of conventional and split approaches. Split-FISH leverages a split-probe design to achieve enhanced specificity for multiplexed FISH applications, which is particularly beneficial when assaying tissue samples. (Copyright: A*STAR’s Genome Institute of Singapore)
SINGAPORE – A*STAR’s Genome Institute of Singapore (GIS) reported a breakthrough probe design for the widely-used biochemical labelling method called fluorescence in situ hybridisation, which is more commonly known as FISH. Conventionally, the FISH technique uses fluorescent DNA probes that bind complementarily to DNA or RNA targets. Improving on previous FISH designs, the team developed split-FISH, a cutting-edge technology that makes it easier to generate highly-detailed maps of gene expressions within cells and tissues. The method could enable massively-multiplexed detection of gene expressions and analysis of gene regulatory networks in tissues. This study was published in Nature Methods on 15 June 2020.
Spatial information is the key to unlocking many biological mysteries, including how cells develop their identities as a result of their unique surroundings in the tissue. Split-FISH is part of a group of new technologies that allows the user to determine unique cellular identities in relation to their location within native tissues. By combining imaging, combinatorial labelling, and clever probe design, Split-FISH maps the molecular details within multiple mouse organs, giving us a glimpse into their inner workings.
Dr Chen Kok Hao, Senior Research Scientist at GIS and corresponding author of this study, commented, “Split-FISH technology could accelerate numerous tissue-based biological research areas, such as neuroscience, developmental biology, immuno-oncology and host-pathogen interactions. Beyond basic science, its deployment to clinical applications, such as pathology and pathogen detection, could also greatly benefit public health.”
GIS and Applied Materials (Applied) have been collaborating on a next-generation histopathology platform to allow spatially-resolved molecular imaging at the subcellular scale. The collaboration combines GIS’ in situ genomic techniques with Applied’s expertise in rapid nanoscale imaging and analysis. While the solution is initially intended for research-use only in immuno-oncology and neurodegenerative diseases, personalised or precision medicine is driving the demand by researchers for new technologies which allow deeper interrogation of intact tissues. The high-resolution, high-throughput solution aims to allow researchers and clinicians to assay individual cells in their native environment, potentially establishing relationships between genotype and phenotype. This could in turn allow the identification of spatial biomarkers and provide clinically actionable insights.
Prof Patrick Tan, Executive Director of GIS, said, “The co-development of the integrated imaging solution could also lead to further innovations in the field of single-cell genomics imaging, and open up new markets that utilise single-cell imaging in a high-throughput manner.”.png?sfvrsn=f4b1bf8d_0)
Gene expression maps of various mouse tissues. Individual RNA molecules of selected genes are depicted as coloured dots. Split-FISH analysis on multiple mouse organs revealed diverse and previously unknown localisation patterns for spatial regulation in mammalian tissues. (Copyright: A*STAR’s Genome Institute of Singapore)
- End -
About A*STAR’s Genome Institute of Singapore (GIS)
The Genome Institute of Singapore (GIS) is an institute of the Agency for Science, Technology and Research (A*STAR). It has a global vision that seeks to use genomic sciences to achieve extraordinary improvements in human health and public prosperity. Established in 2000 as a centre for genomic discovery, the GIS pursues the integration of technology, genetics and biology towards academic, economic and societal impact, with a mission to "read, reveal and write DNA for a better Singapore and world".
Key research areas at the GIS include Precision Medicine & Population Genomics, Genome Informatics, Spatial & Single Cell Systems, Epigenetic & Epitranscriptomic Regulation, Genome Architecture & Design, and Sequencing Platforms. The genomics infrastructure at the GIS is also utilised to train new scientific talent, to function as a bridge for academic and industrial research, and to explore scientific questions of high impact.
For more information about GIS, please visit www.a-star.edu.sg/gis.
About the Agency for Science, Technology and Research (A*STAR)
The Agency for Science, Technology and Research (A*STAR) is Singapore's lead public sector R&D agency, spearheading economic-oriented research to advance scientific discovery and develop innovative technology. Through open innovation, we collaborate with our partners in both the public and private sectors to benefit society.
As a Science and Technology Organisation, A*STAR bridges the gap between academia and industry. Our research creates economic growth and jobs for Singapore, and enhances lives by contributing to societal benefits such as improving outcomes in healthcare, urban living, and sustainability.
We play a key role in nurturing and developing a diversity of talent and leaders in our Agency and research entities, the wider research community and industry. A*STAR’s R&D activities span biomedical sciences and physical sciences and engineering, with research entities primarily located in Biopolis and Fusionopolis. For ongoing news, visit www.a-star.edu.sg.
ANNEX A – NOTES TO EDITOR
The research findings described in this media release can be found in the scientific journal Nature Methods, under the title, “Highly specific multiplexed RNA imaging in tissues with split-FISH” by Jolene Jie Lin Goh1,4, Nigel Chou1,4, Wan Yi Seow1, Norbert Ha1, Chung Pui Paul Cheng1, Yun-Ching Chang2, Ziqing Winston Zhao1,3 & Kok Hao Chen1.
1. Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), Singapore, Singapore.
2. Applied Materials, Inc., Santa Clara, CA, USA.
3. Department of Chemistry and Centre for BioImaging Sciences, National University of Singapore, Singapore, Singapore.
4. These authors contributed equally: Jolene Jie Lin Goh, Nigel Chou.
# Corresponding author: Chen Kok Hao (chenkh@gis.a-star.edu.sg)
Was This Article Helpful?
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
