News
Groundbreaking technology sheds light on the mysteries of RNA modifications

(Standing left) SAM Tsz Wing, Sara (Standing right) Jackie TEO
(Sitting left) Kiyofumi HAMASHIMA (Sitting right) Gary WONG
IMCB scientists have developed an innovative method for rapid capturing of an important type of RNA modification known as N6-methyladenosine (m6A).
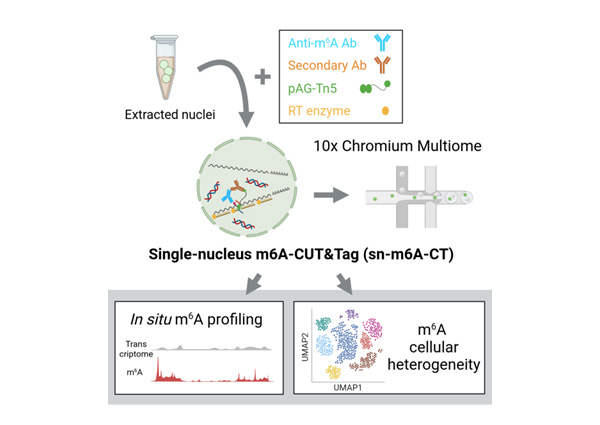
This new technology, known as “single-nucleus m6A-CUT&Tag” (sn-m6A-CT), profiles m6A modifications in unmodified single cells, and is compatible with simultaneous profiling m6A methylomes and transcriptomes in the same cell. This milestone, achieved by the lab of Prof. Jonathan Loh, Deputy Executive Director of IMCB, was recently published in the high-impact journal Molecular Cell.
RNA plays the critical role of carrying genetic information, and its function and behaviour can be tweaked by RNA modifications. m6A, the most abundant modification in mRNA, serves as an adornment on mRNA, orchestrating cellular behaviour and impacting important cellular processes involved in diseases such as cancer, cardiovascular disease, and metabolic conditions. Hence, studying m6A modifications can help us better diagnose and treat diseases.
“Currently, there is no method available for m6A profiling in native populations of cells at the single-cell level,” explained Dr Kiyofumi Hamashima, Senior Scientist in Prof. Loh’s lab and the first author of the study. “Our technology successfully addressed the challenge of signal dilution caused by bulk sampling in a diverse cell population.”
Through this international collaboration involving the Weizmann Institute of Science in Israel and the Mayo Clinic in the USA, Dr. Hamashima and colleagues successfully captured cell-type specific m6A modification landscapes from diverse populations.
In addition, sn-m6A-CT can be combined with existing single-cell transcriptomics methods that analyse the set of RNA molecules in a cell; this can enhance our ability to pinpoint how heterogeneity in RNA modifications affect cell fate identities and states that differ not only by RNA molecules, but also RNA modifications.
“This is the first m6A high-resolution assay in native populations of cells and it has the potential to revolutionise our understanding of RNA heterogeneity and abnormalities,” highlighted Prof. Jonathan Loh, Deputy Executive Director of IMCB.
Importantly, the new technology can provide deeper insights into precision medicine. “These findings provide our researchers and clinicians an immensely valuable platform for investigating molecular signatures from a diagnostic perspective,” said Prof. Hu Li from Mayo Clinic, an international collaborator on this study.
Moving forward, Prof. Loh is excited to use m6A modification information in next-generation diagnostic testing: “Differences in m6A expression has also been associated with disease conditions, suggesting the potential of this technology for future diagnostic applications.” The team is now actively working on integrating microfluidics chips with the technology, with the aim of developing a user-friendly, benchtop-scale, rapid diagnostic device prototype.
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
.png?sfvrsn=c3edc68e_6)