Jonathan LOH Yuin-Han

SUMMARY
KEY AWARDS
- 2023: President's Certificate of Commendation (COVID-19) (For Stronghold Lab)
- 2023: Resilience Medal Award
- 2023: Public Administration Medal (Bronze)
- 2018: NRF Investigatorship Award
- 2017: Outstanding Investigator Award, Stem Cell Society Singapore
- 2015: Ten Outstanding Young Person (TOYP) Award (Scientific and Technological Development),
Junior Camber International (JCI) Singapore - 2012: MIT TR35 Asia Pacific Award
- 2012: World Technology Network Fellowship
- 2011: A*STAR Investigatorship Award
- 2010: Singapore Youth Award
- 2009: Singapore Young Scientist Award
- 2008: Philip Yeo Prize
- 1997: Quest Technology Award
- 2024: Industry Alignment Fund-Prepositioning Project (IAF-PP) (Co-Lead PI) - iPSCs-differentiated Natural Killer cells for cancer immunotherapies (PANAKEIA)
- 2024: Industry Alignment Fund-Prepositioning Project (IAF-PP) (Co-Lead PI) - Engineered extracellular vesicles for anti-cancer therapy (EVANTICA)
- 2023: National Research Foundation-Competitive Research Fund (NRF-CRP) (Lead PI) - Spatial multiome cartography in human thymus to guide nextgen cell based therapies (SPECTRA)
RESEARCH
Research Overview:
Cellular states and fates are influenced by the intricate combinatorial interactions between genetic and epigenetic programs. Our research focuses on mapping and characterizing the diverse potency states of stem cells. To achieve this, we have developed advanced tools and platforms capable of tracing and integrating transcriptomic, epigenomic, and epitranscriptomic information, which helps elucidate the mechanisms regulating cell fate transitions. By leveraging these insights, we aim to engineer specific cell fate functionalities through reprogramming and trans-differentiation, ultimately enhancing our understanding of cellular identity and potential.
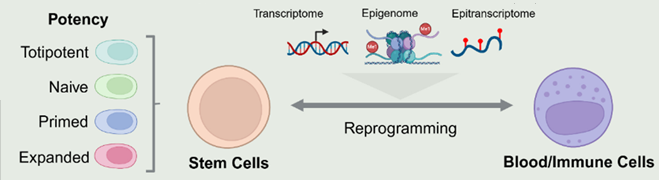 | Schematic of research overview on Cell Fate engineering. Totipotent state - 2-cell embryo stage; primed state - epiblast stage; naive state early blastocyst inner cell mass; expanded state – give rise to germ layers and Trophectoderm (Created using Biorender) |
The first research focus is the intricate relationship between transposable elements (TEs), regulatory factors, and stem cell potency. Our team has identified key factors, including histone chaperones, sumoylation factors, and chromatin modifiers, involved in silencing proviruses and endogenous retroviruses (ERVs) in embryonic stem cells (ESCs), preventing reversion to totipotency. We have also explored the complex epigenetic regulation of TEs, revealing context-specific effects of chromatin modifiers (Cell, 2015). Our work extends to understanding blastoid formation, identifying Nr1h2 as a key regulator influencing both blastoids and blastocysts, with implications for improving embryo models across multiple species (Nat Commun, 2024). Additional studies have explored the roles of ribosomal proteins, ESET (Nucleic Acids Res, 2022), and PRDM15 in stem cell regulation (Nat Genet, 2017), along with the characterization of essential pluripotency enhancers and the metabolic control of stem cell states by Lin28 (Cell Stem Cell, 2016). Collectively, these studies provide significant insights into the mechanisms governing distinct stem cell potency states.
Second research focus is on the cell fate reprogramming and transdifferentiation, demonstrating the amenability of various cell types, including hematopoietic stem and progenitor cells (HSPCs) and T-cells, to reprogramming, while also optimizing the process for efficiency and speed (Blood, 2009 and Cell Stem Cell, 2010). High-throughput screening identified key repressors and effectors influencing reprogramming, with combinatorial knockdown of specific repressors significantly enhancing efficiency. We studied on histone variant H3.3 revealed its bimodal role in maintaining parental cell fate while facilitating transitions to new cell fates (Nat Commun, 2018). Single-cell analyses using scRNA-Seq and scATAC-Seq revealed asynchronous reprogramming trajectories and identified a crucial FOSL1/TEAD4 regulatory network that determines reprogramming success (Sci Adv, 2020). Furthermore, we have developed methods for direct reprogramming of fibroblasts into HSPCs, identifying critical decision points related to cell cycle and competing fate decisions. Finally, our finding in cell differentiation has been applied to enhance the engineering of Natural Killer cells and T cells for immunotherapies, resulting in several intellectual properties licensed to various companies for clinical applications, including GMP-compatible reprogramming, cost-effective cell expansion media, and imprint-free iPSC derivation.
3. Development of a Single-Cell Technologies for Integrating Transcriptome, Epigenome, and Epitranscriptome Information
Last research focus is on developing and applying advanced single-cell multi-omics technologies to investigate cell fate transitions and determination. Our team has pioneered several methods, including a targeted approach combining genotyping with gene expression and DNA methylation analysis, revealing dynamic epigenetic changes during reprogramming. We also developed DARESOME to simultaneously analyze 5mC and 5hmC, highlighting their opposing roles in gene regulation (Sci Adv, 2023). ASTAR-seq allows for simultaneous measurement of transcriptome and chromatin accessibility, enabling the mapping of regulatory landscapes in distinct cell states. Furthermore, our team developed sn-m6A-CT (Mol Cell, 2023), a single-nucleus method for profiling m6A methylomes and transcriptomes, revealing cell-type-specific m6A landscapes and identifying rare cell populations. This method has been further refined to create a comprehensive epitranscriptomic map of mouse post-implantation development, revealing lineage-specific m6A enrichment patterns and the role of m6A demethylase gene in cardiomyocyte maturation. These single-cell platforms have also been instrumental in identifying rare cell populations in the human retina and thymus.
- Nuclear receptor-SINE B1 network modulates expanded pluripotency in blastoids and blastocysts.
Wong KW, Zeng Y, Tay E, Teo JHJ, Cipta NO, Hamashima K, Yi Y, Liu H, Warrier T, Le MTN, Ng SC, Li QJ, Li H, Loh YH.
Nat Commun. 2024 Nov 19;15(1):10011. doi: 10.1038/s41467-024-54381-0. PMID: 39562549 Free PMC article. - Single-nucleus multiomic mapping of m6A methylomes and transcriptomes in native populations of cells with sn-m6A-CT.
Hamashima K, Wong KW, Sam TW, Teo JHJ, Taneja R, Le MTN, Li QJ, Hanna JH, Li H, Loh YH.
Mol Cell. 2023 Aug 25:S1097-2765(23)00649-4. doi: 10.1016/j.molcel.2023.08.010. Online ahead of print.
PMID: 37657444 Free article. - H3.3 safeguards haematopoietic ERV-quilibrium.
Cipta NO, Chen Y, Loh YH.
Nat Cell Biol. 2022 Jan;24(1):7-9. doi: 10.1038/s41556-021-00758-y.
PMID: 34961795 - Diversification of reprogramming trajectories revealed by parallel single-cell transcriptome and chromatin accessibility sequencing.
Xing QR, El Farran CA, Gautam P, Chuah YS, Warrier T, Toh CD, Kang NY, Sugii S, Chang YT, Xu J, Collins JJ, Daley GQ, Li H, Zhang LF, Loh YH.
Sci Adv. 2020 Sep 11;6(37):eaba1190. doi: 10.1126/sciadv.aba1190. Print 2020 Sep.PMID: 32917699 Free PMC article. - Global H3.3 dynamic deposition defines its bimodal role in cell fate transition.
Fang HT, El Farran CA, Xing QR, Zhang LF, Li H, Lim B, Loh YH.
Nat Commun. 2018 Apr 18;9(1):1537. doi: 10.1038/s41467-018-03904-7.
PMID: 29670118 Free PMC article. - Systematic identification of factors for provirus silencing in embryonic stem cells.
Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP, Li Y, Lorincz MC, Tergaonkar V, Lim TM, Chen L, Gunaratne J, Collins JJ, Goff SP, Daley GQ, Li H, Bard FA, Loh YH.
Cell. 2015 Sep 24;163(1):230-45. doi: 10.1016/j.cell.2015.08.037. Epub 2015 Sep 10.PMID: 26365490 Free PMC article. - Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells.
Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, Collins JJ, Daley GQ, Marto JA.
Cell Stem Cell. 2014 Jul 3;15(1):92-101. doi: 10.1016/j.stem.2014.04.002. Epub 2014 May 8.PMID: 24813856 Free PMC article.
- Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients.
Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, Ng HH, Keefe DL, Goldman FD, Klingelhutz AJ, Liu L, Daley GQ.
Nature. 2010 Mar 11;464(7286):292-6. doi: 10.1038/nature08792. Epub 2010 Feb 17.PMID: 20164838 Free PMC article. - Reprogramming of T cells from human peripheral blood.
Loh YH, Hartung O, Li H, Guo C, Sahalie JM, Manos PD, Urbach A, Heffner GC, Grskovic M, Vigneault F, Lensch MW, Park IH, Agarwal S, Church GM, Collins JJ, Irion S, Daley GQ.
Cell Stem Cell. 2010 Jul 2;7(1):15-9. doi: 10.1016/j.stem.2010.06.004.PMID: 20621044 Free PMC article. - The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells.
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH.
Nat Genet. 2006 Apr;38(4):431-40. doi: 10.1038/ng1760. Epub 2006 Mar 5.PMID: 16518401
- A cost-effective and xeno-free medium for human mesenchymal stem cells expansion
A financially efficient and animal component-free culture medium designed to support the robust expansion and proliferation of human mesenchymal stem cells (licensed to company) - A cost-effective, chemically defined medium for human mesenchymal stem cells expansion
A financially sustainable and chemically defined culture medium designed to support the efficient expansion and maintenance of human mesenchymal stem cells while ensuring reproducibility and stability in cell culture conditions (licensed to company)
- Highly efficient method for derivation of genomic imprint-free clinical grade human induced pluripotent stem cells (iPSCs)
A robust and highly efficient approach for generating genomic imprint-free, clinical-grade human induced pluripotent stem cells (iPSCs) with enhanced reliability and suitability for therapeutic applications. - GMP-compatible reprogramming of human blood cells
GMP-compliant methodology for the reprogramming of human blood cells, ensuring safety, reproducibility, and suitability for clinical applications. (licensed to company)
- Method for inducing pluripotency in a hematopoietic cell
A standardized approach for the induction of pluripotency in hematopoietic cells, enabling their reprogramming into a pluripotent state for research and therapeutic applications. (licensed to company)
*Please contact A*STAR if you wish to collaborate or license these technologies.
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
.png?sfvrsn=c3edc68e_6)