Computational Immunology
PI/Head: Mai Chan Lau, Ph.D. (Joint Appointee)
Email: Lau_Mai_Chan@bii.a-star.edu.sg
RSC Website: Computational Immunology on RSC
The Computational Immunology Platform uses state-of-the-art methods to analyse scientific data and deliver pipelines and workflows to accelerate research.
Overview
As a bioinformatics service platform, Computational Immunology provides comprehensive support for diverse data analytics, including exploratory, hypothesis-driven, descriptive, and predictive approaches. Our team
uses state-of-the-art methodologies to deliver pipelines and workflows to
accelerate research. A core strength of the platform is our ability to
integrate analysis across genomics, epigenomics, transcriptomics, proteomics,
metagenomics, as well as single-cell and spatial omics, enabling deeper
insights into complex immunological questions. We work with diverse datasets
including next-generation sequencing (NGS) data, flow cytometry, ELISA,
Quanterix and Luminex assays, long-read sequencing data, histology and
tissue-based molecular imaging data, spatial transcriptomics, with capabilities
to assess cross-correlations across the modalities. Being ISO certified, we adhere
to rigorous standards in the execution of all projects.
Focus Areas
- NGS and NanoString targeted assay analysis (Figure 1)
- Epigenomic Analysis (ChIP-seq and Single-Cell ATAC-Seq) (Figure 2)
- Microbiome analysis (metagenomics and 16s RNA) (Figure 3)
- Spatial transcriptomics analysis (10x Visium, NanoString GeoMx, STOmics) (Figure 4)
- Tissue-based molecular image analysis (IMC data) (Figure 5)
- Integrative analysis (Figure 6)
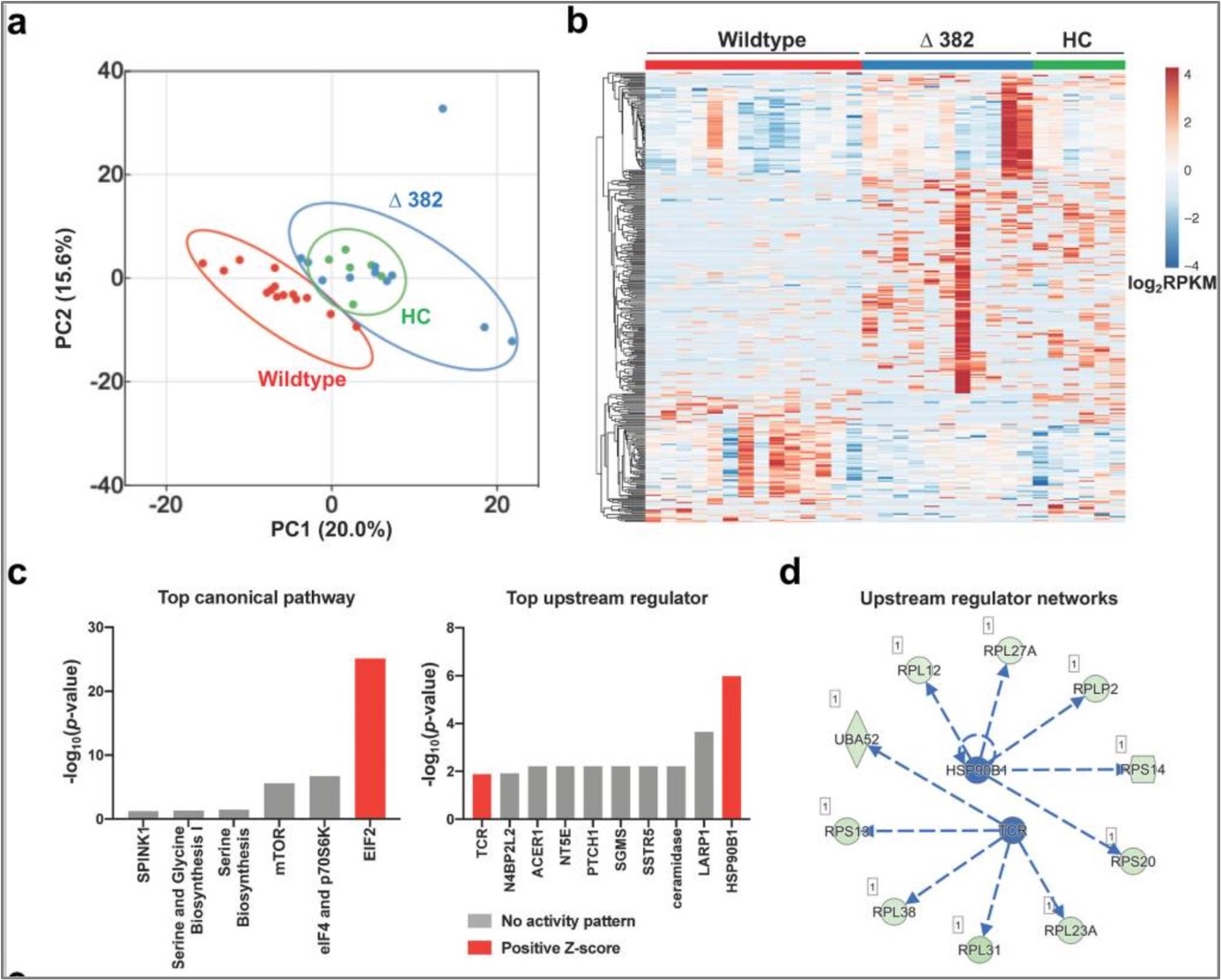
Figure 1: RNAseq analysis of COVID-19 patients and healthy controls (HCs) using (a) principal component analysis, (b) heatmap, (c) top canonical pathways and upstream regulators associated with the DEGs (Ingenuity Pathway Analysis), and (d) integrated network analysis. (1)
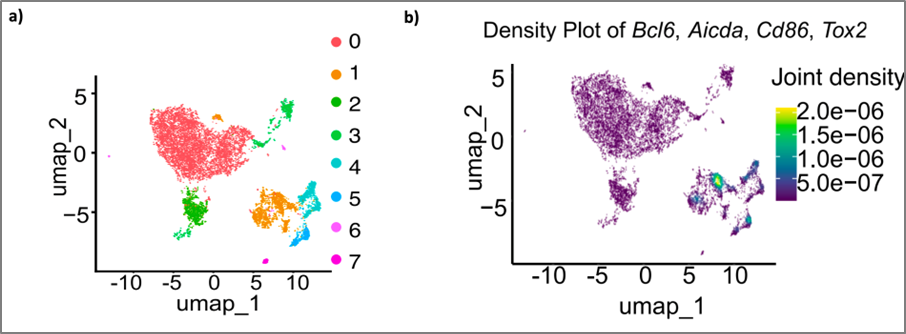
Figure 2: Single-cell ATAC-seq analysis of mouse B cells using (a) UMAP dimensionality reduction to visualize chromatin accessibility across cell populations, (b) Nebulosa density plots to illustrate gene activity patterns (2)
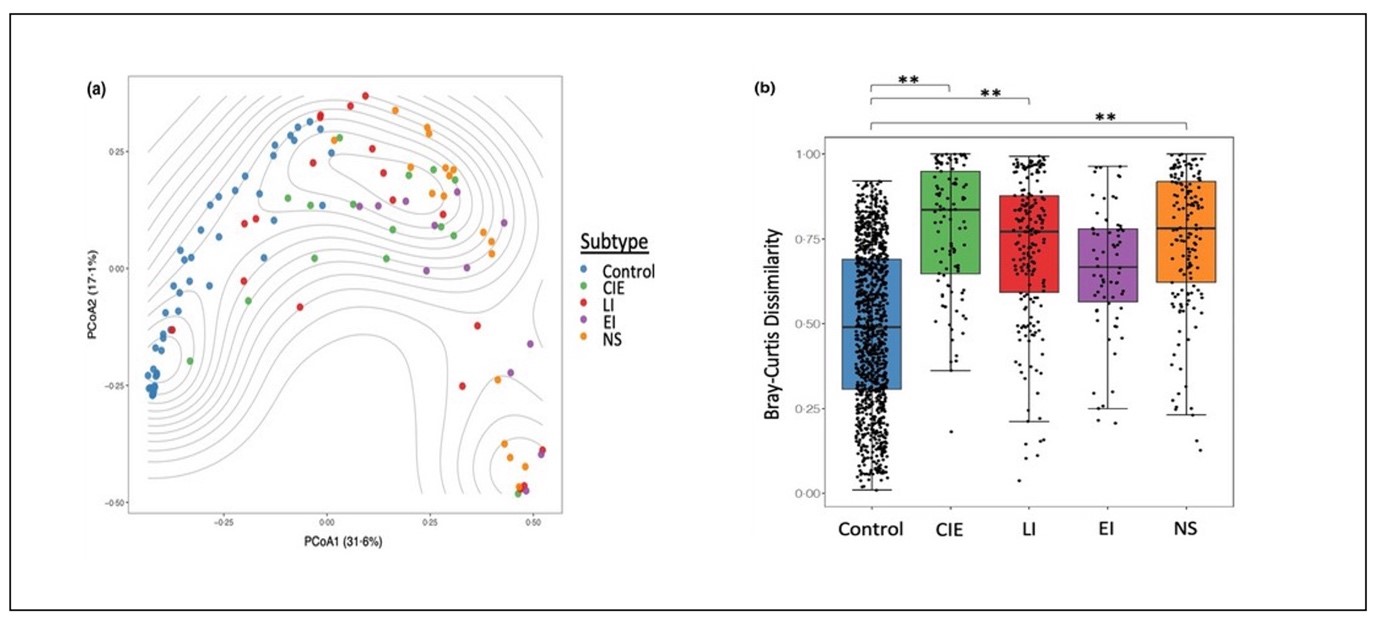
Figure 3: Microbiome analysis of skin metagenomics data using Bray-Curtis dissimilarity (at species-level) visualized in PCoA plot (left) and box plots (right). (3)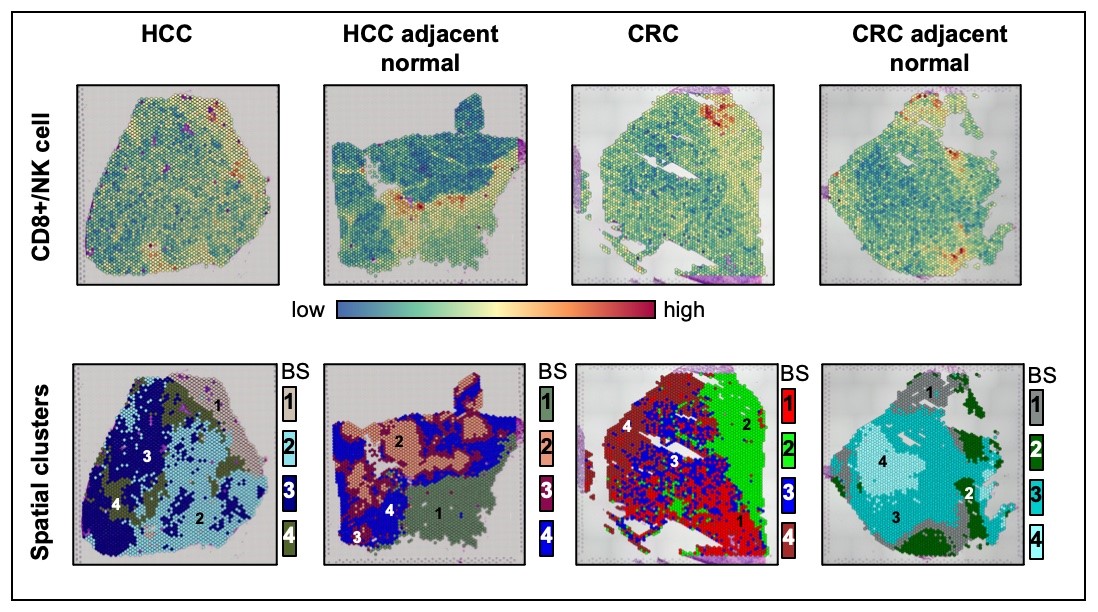
Figure 4: Spatial transcriptomics analysis on SARS-CoV-2 infected tumor samples using Visium (10× Genomics). (4)
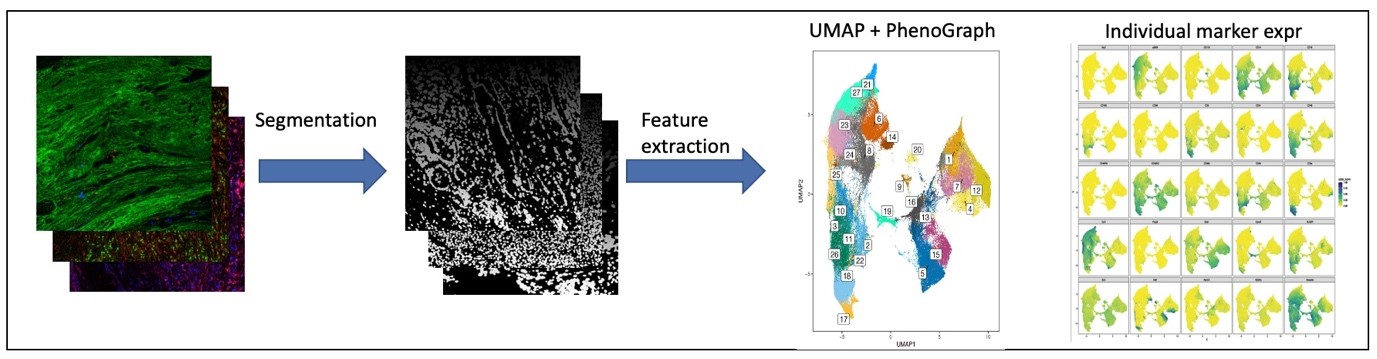
Figure 5: Image processing and downstream cell phenotyping analysis using imaging mass cytometry data.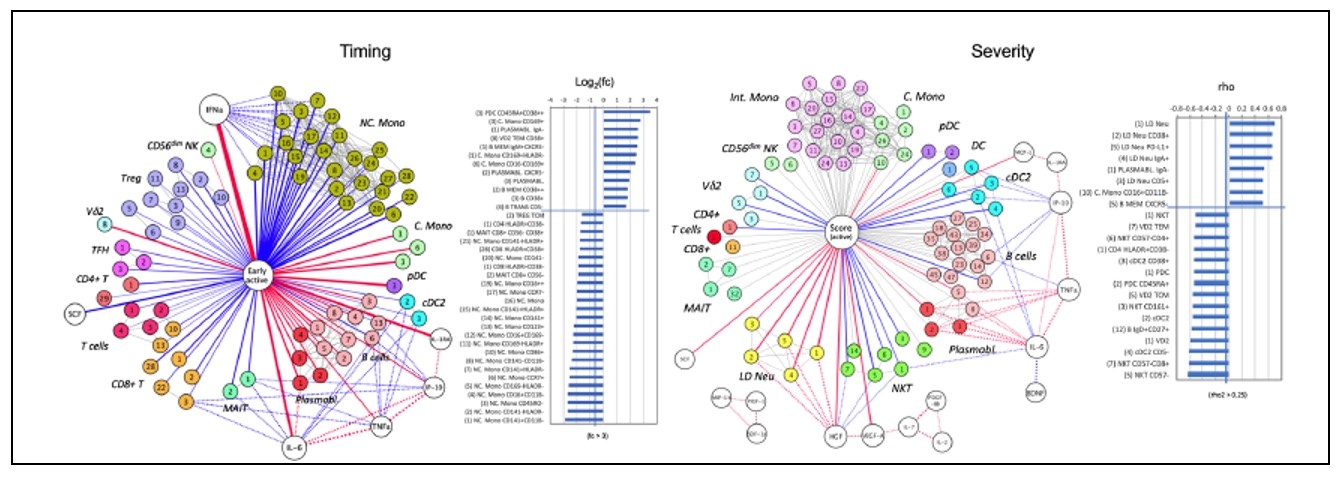
Figure 6: A graph-based network analysis of the interaction between cytokine level and immune cellular frequencies in COVID-19 patients. Association is shown with regard to the timing (left) and the severity (right). (5)
Key Applications & Tools Used
- QIAGEN Ingenuity Pathway Analysis (IPA)
- Tibco Spotfire
- StockinLab (LIMS)
- Blood Requisition System
- Space-Ranger & Cell-Ranger (10X)
- R
- MySQL
- In-House Bioinformatics Tools
- iCellSight (6), HepatoMap (7), and TIPC (8)
Computing / Data Resources
- Multiple Computational Servers for Data Analysis (128+ Core CPUs, 2+TB RAM, 8 high-end GPUs, etc.)
- Cumulative Storage of about 2PB (2000TB)
Selected Publications:
- Fong SW, Yeo NK, Chan YH, Goh YS, Amrun SN, Ang N, Rajapakse MP, Lum J, Foo S, Lee CY, Carissimo G, Chee RS, Torres-Ruesta A, Tay MZ, Chang ZW, Poh CM, Young BE, Tambyah PA, Kalimuddin S, Leo YS, Lye DC, Lee B, Biswas S, Howland SW, Renia L, Ng LFP. Robust Virus-Specific Adaptive Immunity in COVID-19 Patients with SARS-CoV-2 Δ382 Variant Infection. J Clin Immunol. 2022 Feb;42(2):214-229. doi: 10.1007/s10875-021-01142-z. Epub 2021 Oct 30. Erratum in: J Clin Immunol. 2021 Nov 27;: PMID: 34716845; PMCID: PMC8556776.
- Morgan D, Zhang B, Fidan K, Pan W, Vaiyapuri TS, Raju A, Hei D, See TY, Sari IN, Chan JW, Majee P, Balachander A, Yu J, Wong AH, Mok MMH, Foo SH, Tang WL, Ang N, Tan I, Peng YF, Jaynes P, Xu S, Ghosh G, Shahabi S, Jeyasekharan AD, Ikawa M, Zhang Y, Howland SW, Lau MC, Wang VY, Lam KP, Tergaonkar V. The transcription complex p52-ETS1 is essential for germinal center formation. Nat Immunol. 2025 Jul 25. doi: 10.1038/s41590-025-02236-1. Epub ahead of print. PMID: 40715659.
- Tham KC, Lefferdink R, Duan K, Lim SS, Wong XFCC, Ibler E, Wu B, Abu-Zayed H, Rangel SM, Del Duca E, Chowdhury M, Chima M, Kim HJ, Lee B, Guttman-Yassky E, Paller AS, Common JEA. Distinct skin microbiome community structures in congenital ichthyosis. Br J Dermatol. 2022 Oct;187(4):557-570. doi: 10.1111/bjd.21687. Epub 2022 Jul 4. PMID: 35633118; PMCID: PMC10234690.
- Lau MC, Yi Y, Goh D, Cheung CCL, Tan B, Lim JCT, Joseph CR, Wee F, Lee JN, Lim X, Lim CJ, Leow WQ, Lee JY, Ng CCY, Bashiri H, Cheow PC, Chan CY, Koh YX, Tan TT, Kalimuddin S, Tai WMD, Ng JL, Low JG, Lim TKH, Liu J, Yeong JPS. Case report: Understanding the impact of persistent tissue-localization of SARS-CoV-2 on immune response activity via spatial transcriptomic analysis of two cancer patients with COVID-19 co-morbidity. Front Immunol. 2022 Sep 12;13:978760. doi: 10.3389/fimmu.2022.978760. PMID: 36172383; PMCID: PMC9510984.
- Lim J, Puan KJ, Wang LW, Teng KWW, Loh CY, Tan KP, Carissimo G, Chan YH, Poh CM, Lee CY, Fong SW, Yeo NK, Chee RS, Amrun SN, Chang ZW, Tay MZ, Torres-Ruesta A, Leo Fernandez N, How W, Andiappan AK, Lee W, Duan K, Tan SY, Yan G, Kalimuddin S, Lye DC, Leo YS, Ong SWX, Young BE, Renia L, Ng LFP, Lee B, Rötzschke O. Data-Driven Analysis of COVID-19 Reveals Persistent Immune Abnormalities in Convalescent Severe Individuals. Front Immunol. 2021 Nov 19;12:710217. doi: 10.3389/fimmu.2021.710217. PMID: 34867943; PMCID: PMC8640498.
- Yancun Zhu, Felicia Wee, Menaka Priyadharsani Rajapakse, Esther Gek Teo, Nicholas Ang, Solomonraj Wilson, Zheng Yi Ho, Willa Wen-You Yim, Li Yen Chong, Craig Ryan Joseph, Jeffrey Chun Lim, Zhen Wei Neo, Chwee Ming Lim, Bernett Lee, Olaf Rotzschke, Joe Yeong, Mai Chan Lau; Abstract LB252: iCellSight: An interactive tool for analyzing and visualizing in-situ high-plex cellular protein data. Cancer Res 1 April 2024; 84 (7_Supplement): LB252.
- Wei Lin Tang, Teresa J. Tye, Menaka Priyadharsani Rajapakse, Solomonraj Wilson, Olaf RÖtzschke, David Tai, Darren Wan-Teck Lim, Juha P. Väyrynen, Joe Poh Sheng Yeong, Mai Chan Lau; Abstract 5063: Hepatomap: An interactive shiny app for spatial transcriptomics insights in hepatocellular carcinoma. Cancer Res 15 April 2025; 85 (8_Supplement_1): 5063.
- Mai Chan Lau, Jennifer Borowsky, Juha P Väyrynen, Koichiro Haruki, Melissa Zhao, Andressa Dias Costa, Simeng Gu, Annacarolina da Silva, Tomotaka Ugai, Kota Arima, Minh N Nguyen, Yasutoshi Takashima, Joe Yeong, David Tai, Tsuyoshi Hamada, Jochen K Lennerz, Charles S Fuchs, Catherine J Wu, Jeffrey A Meyerhardt, Shuji Ogino, Jonathan A Nowak. Tumor-immune partitioning and clustering algorithm for identifying tumor-immune cell spatial interaction signatures within the tumor microenvironment. PLoS Comput Biol. 2025 Feb 18;21(2):e1012707. doi: 10.1371/journal.pcbi.1012707. PMID: 39965007 PMCID: PMC11849983.
- Tay RE, Ho CM, Ang NDZ, Tay HC, Lopez DZ, Na QR, Tan YW, Koh SM, Tan KP, Lee W, Lim J, Lau MC, Toh HC, Rotzschke O, Rénia L. Serotonin receptor 5-HT2A as a potential target for HCC immunotherapy. JITC. 2025;13:e011088.
- Neo SY, Shuen TWH, Khare S, Chong J, Lau M, Shirgaonkar N, Chua L, Zhao J, Lee K, Tan C, Ba R, Lim J, Chua J, Cheong HS, Chai HM, Chan CY, Chung AYF, Cheow PC, Jeyaraj PR, Teo JY, Koh YX, Chok AY, Chow PKH, Goh B, Wan WK, Leow WQ, Loh TJZ, Tang PY, Karunanithi J, Ngo NT, Lim TKH, Xu S, Dasgupta R, Toh HC, Lam KP. Atypical memory B cells acquire Breg phenotypes in hepatocellular carcinoma. JCI Insight. 2025 Feb 25:e187025. doi: 10.1172/jci.insight.187025. Epub ahead of print. PMID: 39998891.
- Lau MC, Borowsky J, Väyrynen JP, Haruki K, Zhao M, Dias Costa A, Gu S, da Silva A, Ugai T, Arima K, Nguyen MN, Takashima Y, Yeong J, Tai D, Hamada T, Lennerz JK, Fuchs CS, Wu CJ, Meyerhardt JA, Ogino S, Nowak JA. Tumor-immune partitioning and clustering algorithm for identifying tumor-immune cell spatial interaction signatures within the tumor microenvironment. PLoS Comput Biol. 2025 Feb 18;21(2):e1012707. doi: 10.1371/journal.pcbi.1012707. PMID: 39965007; PMCID: PMC11849983.
- Ugai T, Väyrynen JP, Ugai S, Zhong R, Haruki K, Lau MC, Zhao M, Zhong Y, Yao Q, Matsuda K, Guerriero JL. Long-term marine ω-3 polyunsaturated fatty acids intake in relation to incidence of colorectal cancer subclassified by macrophage infiltrates. The Innovation Medicine. 2024 Aug 17;2(3):100082-. Doi: 10.59717/j.xinn-med.2024.100082
- Azam AB, Wee F, Väyrynen JP, Yim WWY, Xue YZ, Chua BL, Lim JCT, Somasundaram AC, Tan DSW, Takano A, Chow CY, Khor LY, Lim TKH, Yeong J, Lau MC, Cai Y. Training immunophenotyping deep learning models with the same-section ground truth cell label derivation method improves virtual staining accuracy. Front Immunol. 2024 Jun 28;15:1404640.
- Zhang Y, Lee RY, Tan CW, Guo X, Yim WWY, Lim JCT, Wee FYT, Wu Y, Kharbanda M, Lee JYJ, Ngo NT, Leow WQ, Loo LH, Lim TKH, Sobota RM, Lau MC, Davis MJ, Yeong J. Spatial omics techniques and data analysis for cancer immunotherapy applications. Curr Opin Biotechnol. 2024: 103111. ISSN 0958-1669. doi:10.1016/j.copbio.2024.103111.
- Elomaa H, Härkönen J, Väyrynen SA, Ahtiainen M, Ogino S, Nowak JA, Lau MC, Helminen O, Wirta EV, Seppälä TT, Böhm J, Mecklin JP, Kuopio T, Väyrynen JP. Quantitative multiplexed analysis of IDO and ARG1 expression and myeloid cell infiltration in colorectal cancer. Mod Pathol. 2024 Feb 16:100450. doi: 10.1016/j.modpat.2024.100450. Epub ahead of print. PMID: 38369188.
- Fok-Moon Lum, Yi-Hao Chan, Teck-Hui Teo, Etienne Becht, Siti Naqiah Amrun, Karen WW Teng, Siddesh V Hartimath, Nicholas KW Yeo, Wearn-Xin Yee, Nicholas Ang, Anthony M Torres-Ruesta, Siew-Wai Fong, Julian L Goggi, Evan W Newell, Laurent Renia, Guillaume Carissimo, and Lisa FP Ng .Crosstalk between CD64+MHCII+ macrophages and CD4+ T cells drives joint pathology during chikungunya. EMBO Molecular Medicine (2024). 10.1038/s44321-024-00028-y.
- Meng J, Tan JYT, Joseph CR, Ye J, Lim JCT, Goh D, Xue Y, Lim X, Koh VCY, Wee F, Tay TKY, Chan JY, Ng CCY, Iqbal J, Lau MC, Lim EH, Toh HC, Teh BT, Dent RA, Tan PH, Joe Yeong YPS. The prognostic value of CD39 as a marker of tumor-specific T cells in triple-negative breast cancer in Asian women. Lab. Invest. 2023. 100303. ISSN 0023-6837. https://doi.org/10.1016/j.labinv.2023.100303.
- Hong JH, Yong CH, Heng HL, Chan JY, Lau MC, Chen J, Lee JY, Lim AH, Li Z, Guan P, Chu PL, Boot A, Ng SR, Yao X, Wee FYT, Lim JCT, Liu W, Wang P, Xiao R, Zeng X, Sun Y, Koh J, Kwek XY, Ng CCY, Klanrit P, Zhang Y, Lai J, Tai DWM, Pairojkul C, Dima S, Popescu I, Hsieh S-Y, Yu M-C, Yeong J, Kongpetch S, Jusakul A, Loilome W, Tan P, Tan J, The BT. Integrative multiomics enhancer activity profiling identifies therapeutic vulnerabilities in cholangiocarcinoma of different etiologies. Gut. Published Online First: 24 November 2023. doi: 10.1136/gutjnl-2023-330483.
- Cong, SL., Bok, LC., Wee F., Ng CW, Ang, NDZ, Sim TJR, Yuan, C., Rajapakse, MP, Wilson, S., Lim, J., Azam, ABB, Cai Y., Yeong, J., Lau MC. 1279 H&E-2.0: an extended application of ST-net to liver cancer and its generalizability. Journal for ImmunoTherapy of Cancer 11(Suppl 1): 2023, A1417-A1419.
- Feng, X., Zhang, M., Yim, W., Wee, F., Rajapakse, MP., Lim, J., Ng, CW., Zlobec, I., Lee, B., Rotzchke, O., Yeong, J., Lau, MC. 1282 H&E 2.0 virtual staining of CD3 T cells: improving cell labeling for precise model training. Journal for ImmunoTherapy of Cancer 11(Suppl 1): 2023, A1422-A1423.
- Kaya NA, Tai D, Lim X, Lim JQ, Lau MC, Goh D, Phua CZJ, Wee FYT, Joseph CR, Lim JCT, Neo ZW, Ye J, Cheung L, Lee J, Loke KSH, Gogna A, Yao F, Lee MY, Shuen TWH, Toh HC, Hilmer A, Chan YS, Lim TKH, Tam WL, Choo SP, Joe Y, Zhai W. Multimodal molecular landscape of response to Y90-resin microsphere radioembolization followed by nivolumab for advanced hepatocellular carcinoma. JITC. 2023;11:e007106. doi: 10.1136/jitc-2023-007106.
- Cheung CCL, Seah YHJ, Fang J, Orpilla NHC, Lau MC, Lim CJ, Lim X, Lee JNLW, Lim JCT, Lim S, Cheng Q, Toh HC, Choo SP, Lee SY, Lee JJX, Liu J, Lim TKH, Tai D and Yeong J (2023) Immunohistochemical scoring of LAG-3 in conjunction with CD8 in the tumor microenvironment predicts response to immunotherapy in hepatocellular carcinoma. Front. Immunol. 14:1150985. doi: 10.3389/fimmu.2023.1150985.
- Lee RY, Ng CW, Rajapakse MP, Ang N, Yeong J, Lau MC. The promise and challenge of spatial omics in dissecting tumour microenvironment and the role of AI. Front Oncol, 13, 1837.
- Lee R, Wu Y, Goh D, Tan V, Ng CW, Lim JC, Lau MC, Sheng JY. Application of Artificial Intelligence to In Vitro Tumor Modeling and Characterization of the Tumor Microenvironment. Adv. Healthc. Mater. 2023 Apr 15:2202457.
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
