Detecting a wide range of epitranscriptomic modifications using a nanopore-sequencing-based computational approach with 1D score-clustering
10 December 2024 - Over the past few decades, it has become increasingly clear that epigenetic and epitranscriptomic modifications are crucial in regulating a wide range of functions across all forms of life. These modifications affect major biological processes, including those responsible for transcription, pre-messenger RNA (pre-mRNA) splicing, nuclear export, translation, cellular differentiation, mRNA stability, immune cell biology, neuronal development and brain plasticity, depression, embryonic development, viral lifecycle, inflammation, cardiovascular disease, cancer, aging, and others.
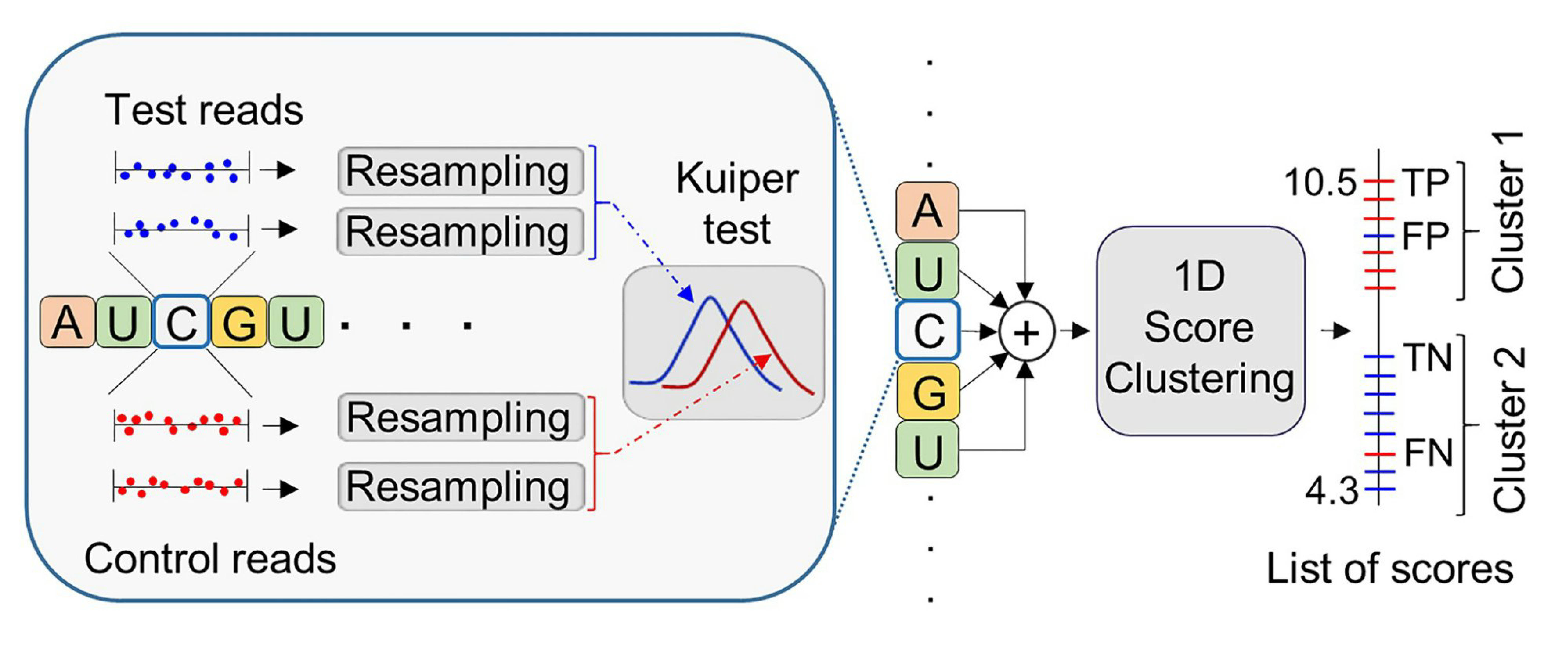
To date, over 40 types of DNA and 300 types of RNA modifications have been identified, but current methods can detect less than 10% of these. Long-read sequencing and advanced computational techniques, like machine learning, offer a solution. Modena, a new unsupervised learning method using long-read nanopore sequencing, can detect many modifications. Developed by the team at GIS - Mile Sikic, Jianjun (JJ) Liu, Sara Bakić, Zhe Li and Prof Roger Foo from NUS, Modena uses ‘dynamic thresholding,’ a unique method that improves performance significantly when combined with other algorithms. It outperformed other methods in most tests on RNA and showed consistent accuracy on a DNA dataset. Read more about their latest findings at https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkae1168/7919996?utm_source=advanceaccess&utm_campaign=nar&utm_medium=email discovery of a new clade of Candida Auris.
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
.png?sfvrsn=2e525642_5)