DUBStepR
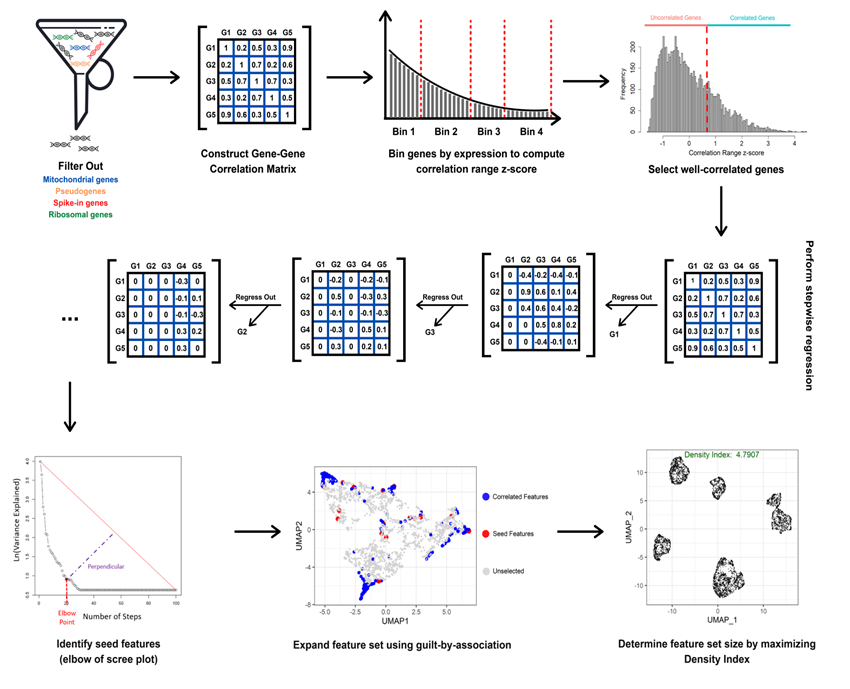
| Name of software | DUBStepR(Determining the Underlying Basis using Stepwise Regression) |
| Purpose | DUBStepR is an algorithm for feature selection based on gene–gene correlations |
| Name of Contact | Ignasius Joanito & Shyam Prabhakar |
| Email of technical contact | Ignasius_joanito_irwan@gis.a-star.edu.sg prabhakars@gis.a-star.edu.sg |
| Summary of software function | DUBStepR is an algorithm for feature selection based on gene–gene correlations. A key feature of DUBStepR is the use of a stepwise approach to identify an initial core set of genes that most strongly represent coherent expression variation in the dataset. Uniquely, DUBStepR defines a novel graph-based measure of cell aggregation in the feature space (termed density index (DI)), and uses this measure to optimize the number of features. DUBStepR requires R version >= 3.5.0. DUBStepR is available as an R package on CRAN (https://CRAN.R-project.org/package=DUBStepR), and is well documented for easy integration into the Seurat pipeline. The source code is also freely available on GitHub (https://github.com/prabhakarlab/DUBStepR). |
| Publications describing software & its application | Ranjan, B., Sun, W., Park, J. et al. DUBStepR is a scalable correlation-based feature selection method for accurately clustering single-cell data. Nat Commun 12, 5849 (2021). |
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
.png?sfvrsn=2e525642_5)