GIS RESEARCHERS COLLABORATE WITH CLINICIANS TO REVIEW ROLE OF SKIN MICROBIOME IN ECZEMA
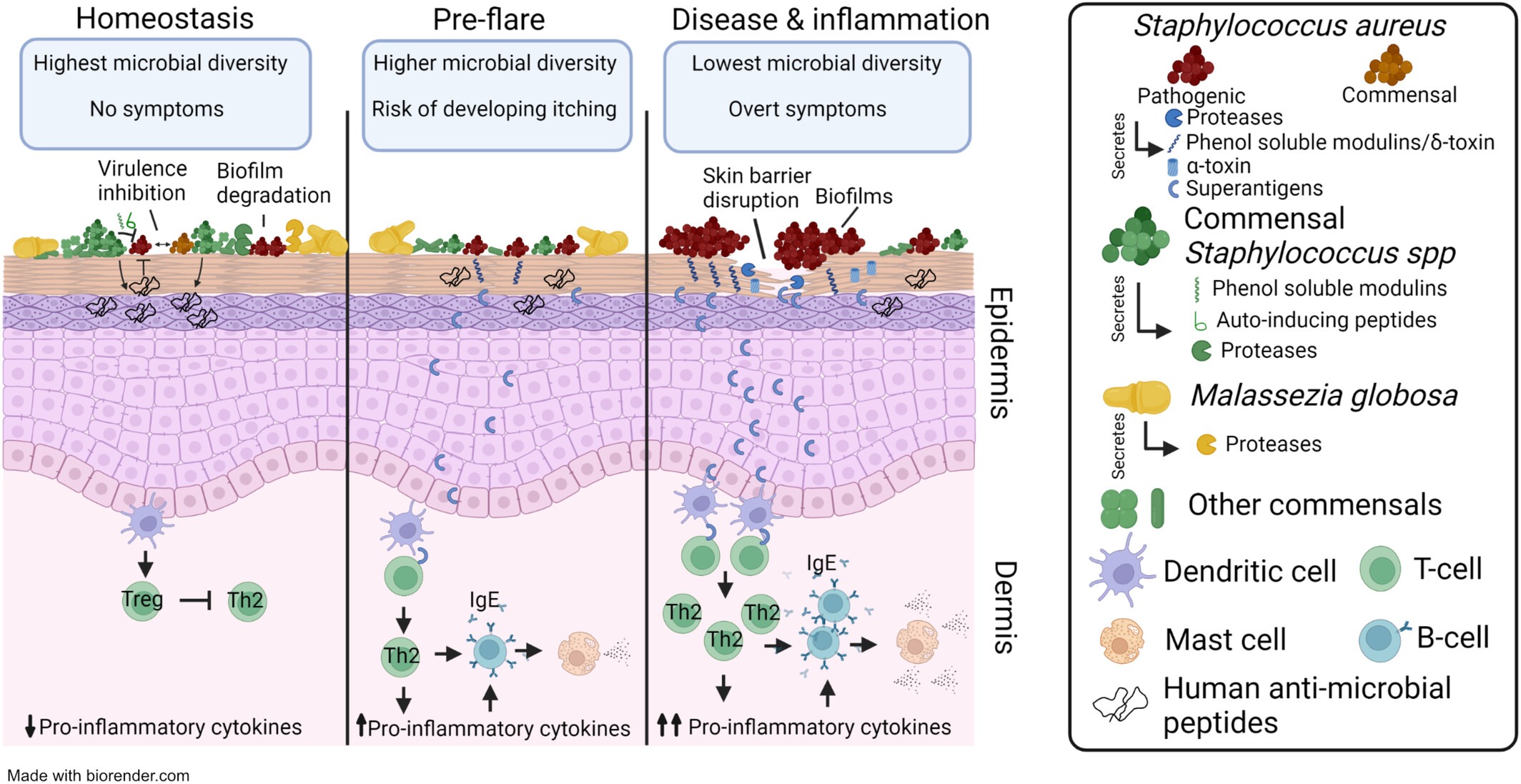
Source: Allergy (https://doi.org/10.1111/all.16044)
3 February 2024 - Atopic dermatitis (eczema) is a common recurring skin condition affecting 25% of children and has a substantial impact on their quality of life due to highly visible flares, itching and a loss of self-esteem. Eczema is often subsequently accompanied by other chronic conditions such as food allergies and auto-immune diseases. Thus, there is great clinical interest in early prevention of eczema in children. However, eczema is a clinically heterogenous disease and flares can be triggered or exacerbated by multiple factors such as patient genetics, irritants from the environment or even the bacteria that reside on our skin.
Dr Niranjan Nagarajan, Principal Investigator at A*STAR’s Genome Institute of Singapore (GIS) and Dr Minghao Chia, GIS fellow, collaborated with clinicians from the National University Health System (NUHS) of Singapore and other domain experts from A*STAR Skin Research Labs (A* SRL) and National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health (NIH) in the United States to author a review about the complex interplay between human genetics, the immune system and the skin microbiome in the aetiology of AD and food allergies.
In this review “The skin microbiome in pediatric atopic dermatitis and food allergy” published in Allergy, the team summarized the current scientific understanding of the role of the skin microbiome in eczema. Topics covered include the possible molecular mechanisms by which microbes worsen or protect against eczema and food allergies, how early life (< 1 year) skin microbial signatures may be predictive of future disease, how household transmission of microbes may contribute to eczema recurrence, and how immunotherapies can shape the skin microbiome. Importantly, many of these findings were made possible due to advances in genomic technologies. The authors also identified several promising clinical trials harnessing the skin microbiome for targeted treatment of eczema and food allergies such as the topical application of protective microbes or their products.
Nagarajan and Chia believe that state-of-the-art genomic technologies can help with dissecting the factors that contribute to eczema. “Technologies like Nanopore long read sequencing, metagenomics (DNA seq) and metatranscriptomics (RNA seq) can identify eczema-associated human and microbial pathways or products which can vary across patients. This is crucial for better patient stratification and targeted treatment,” said the researchers from GIS.
The authors also highlighted gaps in existing literature that should be prioritized in future work. First, researchers need to determine whether changes in the skin microbiome can influence eczema severity or are just signs of disease or treatment. Second, the integration of genomics, microbial culture-based experiments and host measurements are needed for a mechanistic understanding of the role these factors play in disease. Third, time-course studies are required to pinpoint the best time for intervention for greatest clinical efficacy. Fourth, more clinical trials should be conducted to assess how host and extrinsic factors modify the effectiveness of microbe-based therapies for managing eczema and food allergies.
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM
.png?sfvrsn=2e525642_5)