Function and Structure of RNA

Huber Roland G.
 | HUBER Roland G. Principal Investigator Email: rghuber@bii.a-star.edu.sg Research Group: Function and Structure of RNA |
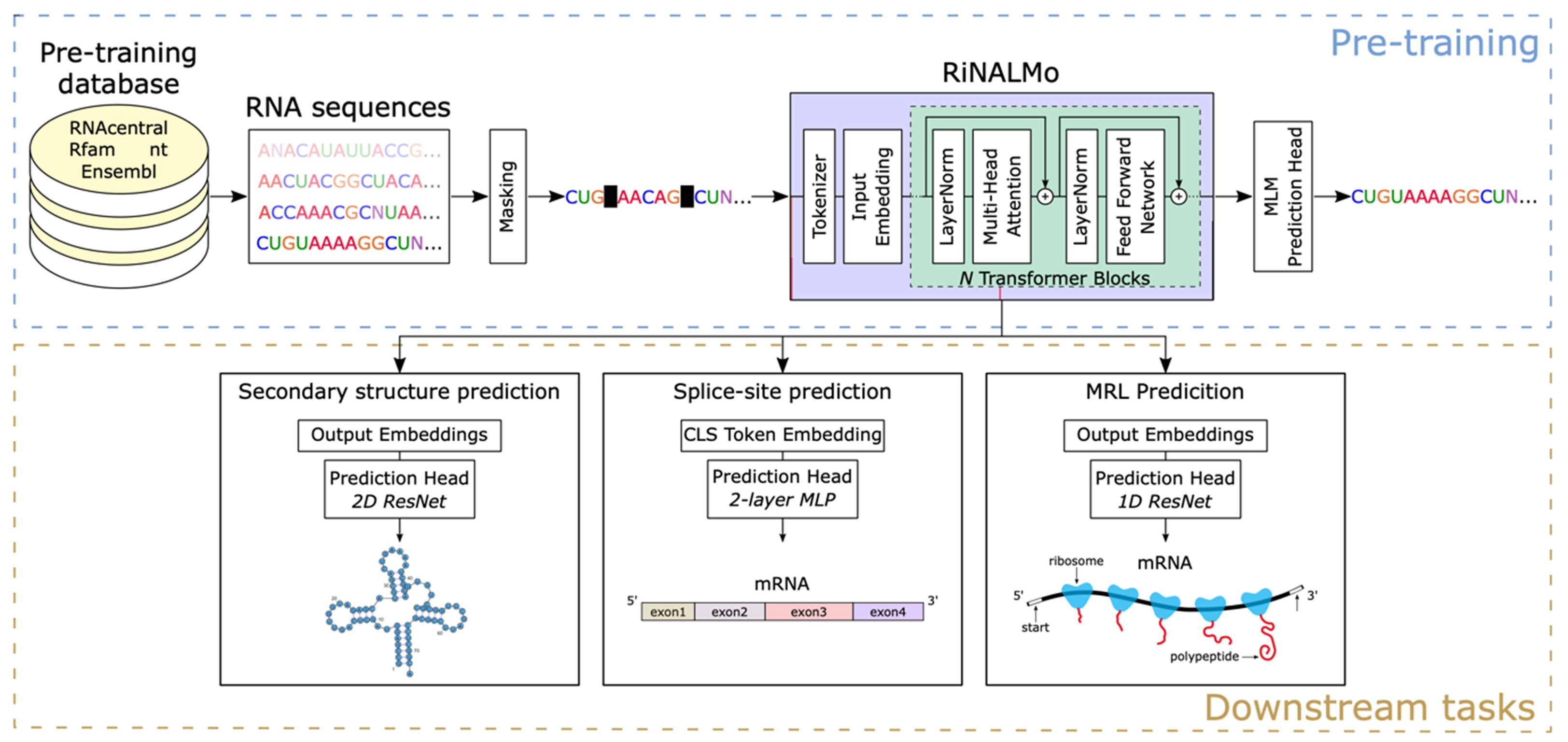
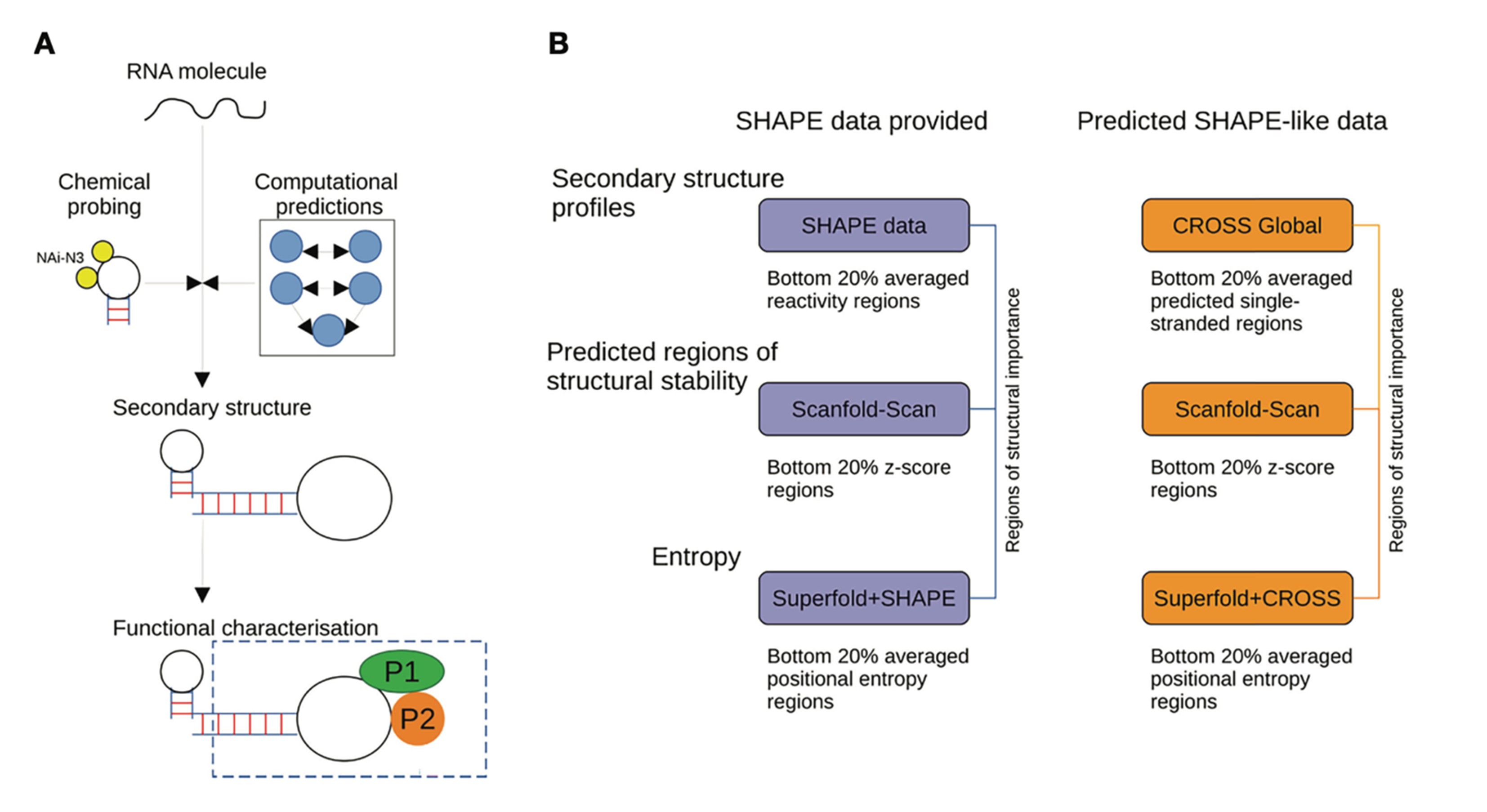
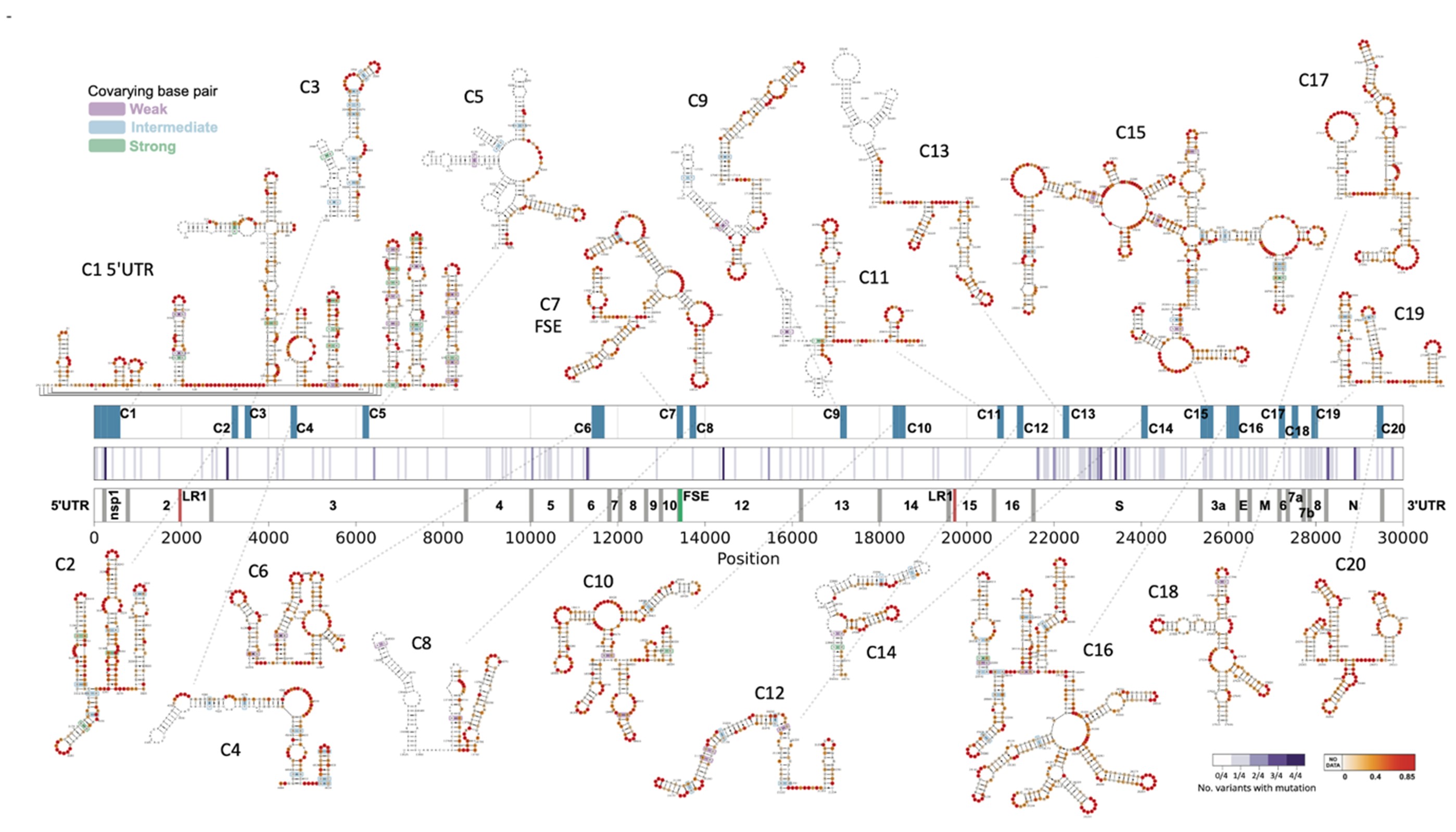
Dr. Roland G. Huber studied computational biology at the University of Innsbruck and at Yale University. After obtaining his PhD he commenced postdoctoral research at A*STAR in Singapore and was appointed as principal investigator in 2019. His group at the Bioinformatics Institute is primarily interested in the interplay of structure and function of RNA in gene regulation and infectious disease using statistical modeling, multi-omics data, and molecular simulations to reveal the structures and functional mechanisms of folded RNA and RNA-protein complexes. The group has previously worked on a variety of tropical and emerging infectious diseases including Dengue, Zika, and SARS-CoV-2. Other areas of interest are multi-omics and metagenomic analysis, particularly in the fields of animal health & aquaculture, and human immunology & ageing
Group Member(s)
| Senior Scientist | DEFALCO Louis |
| Senior Scientist | KULKARNI Mandar |
| Research Officer | CHIAM Aryeh Joseph |
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM