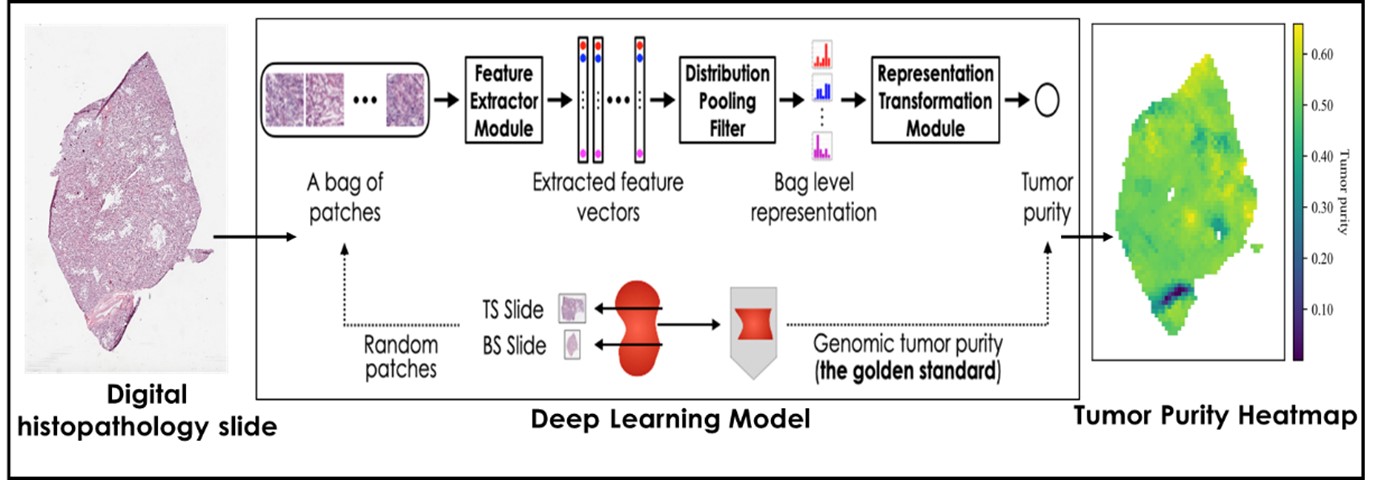
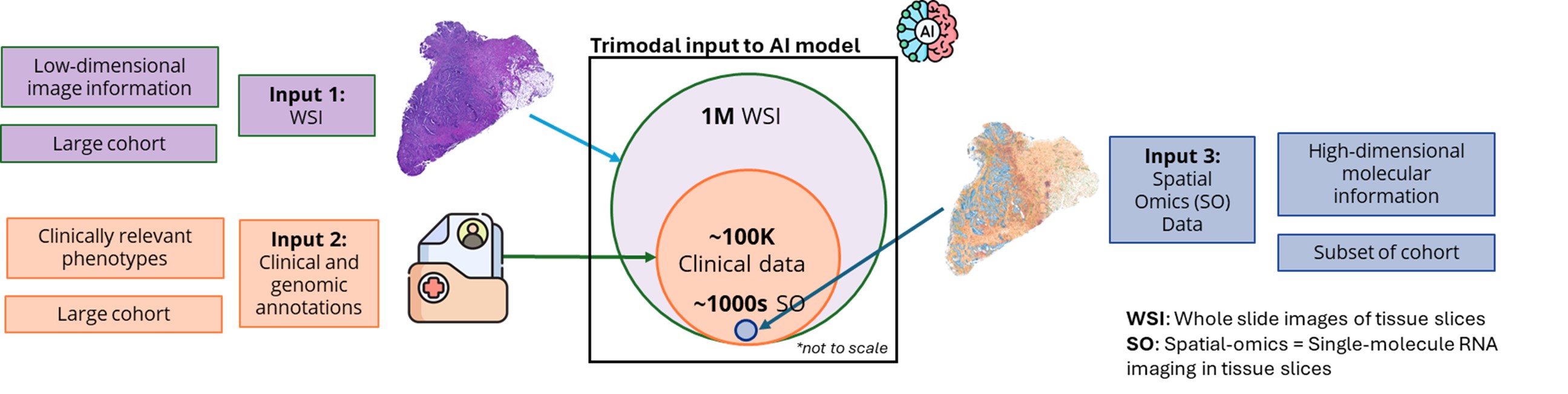
Computer Vision and Pattern Discovery

Lee Hwee Kuan
 | LEE Hwee Kuan (Head) Deputy Director (Training and Talent) / Senior Principal Scientist Email: leehk@a-star.edu.sg Research Group: Computer Vision and Pattern Discovery |
Dr. Lee Hwee Kuan is a Senior Principal Investigator of the Imaging Informatics division in Bioinformatics Institute. His current research work involves developing of computer vision aglorithms for clinical and biological studies. Dr. Lee obtained his Ph.D. in 2001 in Theoretical Physics from Carnegie Mellon University with a thesis on liquid-liquid phase transitions and quasicrystals. He then held a joint postdoctoral position with Oak Ridge National Laboratory (USA) and University of Georgia where he worked on developing advanced Monte Carlo methods and nano-magnetism. In 2003, with an award from the Japan Society for Promotion of Science, Hwee Kuan moved to Tokyo Metropolitan University where he developed solutions to extremely long time scaled problems and a reweighting method for nonequilibrium systems. In 2005, Dr. Lee returned to Singapore and join Data Storage Institute, A*STAR on investigating novel recording methods such as hard disk recording via magnetic resonance. In 2006, Dr. Lee transferred to BII, A*STAR as Principal Investigator and he leaded the Imaging Informatics Division.
Group Members
| Senior Scientist | LIU Wei |
| Senior Scientist | SINGH Malay |
| Post Doc Research Fellow (Collaborator) | PARK Sojeong |
| Lead Research Officer (T-UP) | SRINIVASA Channarayapatna Arvind |
| Senior Research Officer | GOH Jie Hui Corinna |
| Research Officer | ZHANG Tianyi |
| Research Officer | CHOO Yu Liang |
| Research Officer | SHIMAZAWA Kei |
| Research Officer | HESTER James |
| PhD Student | REN Yu |
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM