Computational Chemical Biology and Fragment-Based Design

Tan Yaw Sing
 | TAN Yaw Sing Principal Investigator Email: tan_yaw_sing@a-star.edu.sg Research Group: Computational Chemical Biology and Fragment-Based Design |
Dr. Tan Yaw Sing graduated with a B.Sc. (Hons) from the National University of Singapore in 2009, with a double major in Chemistry and Life Sciences. Funded by the A*STAR Graduate Scholarship, he proceeded to pursue his postgraduate studies at the University of Cambridge, where he obtained his PhD in Chemistry in 2014. In the same year, he joined Dr. Chandra Verma’s group at the Bioinformatics Institute (BII) as a postdoctoral research fellow. He has been leading his own group in BII since April 2021 and has been appointed as Principal Investigator in April 2024.
Research Interests
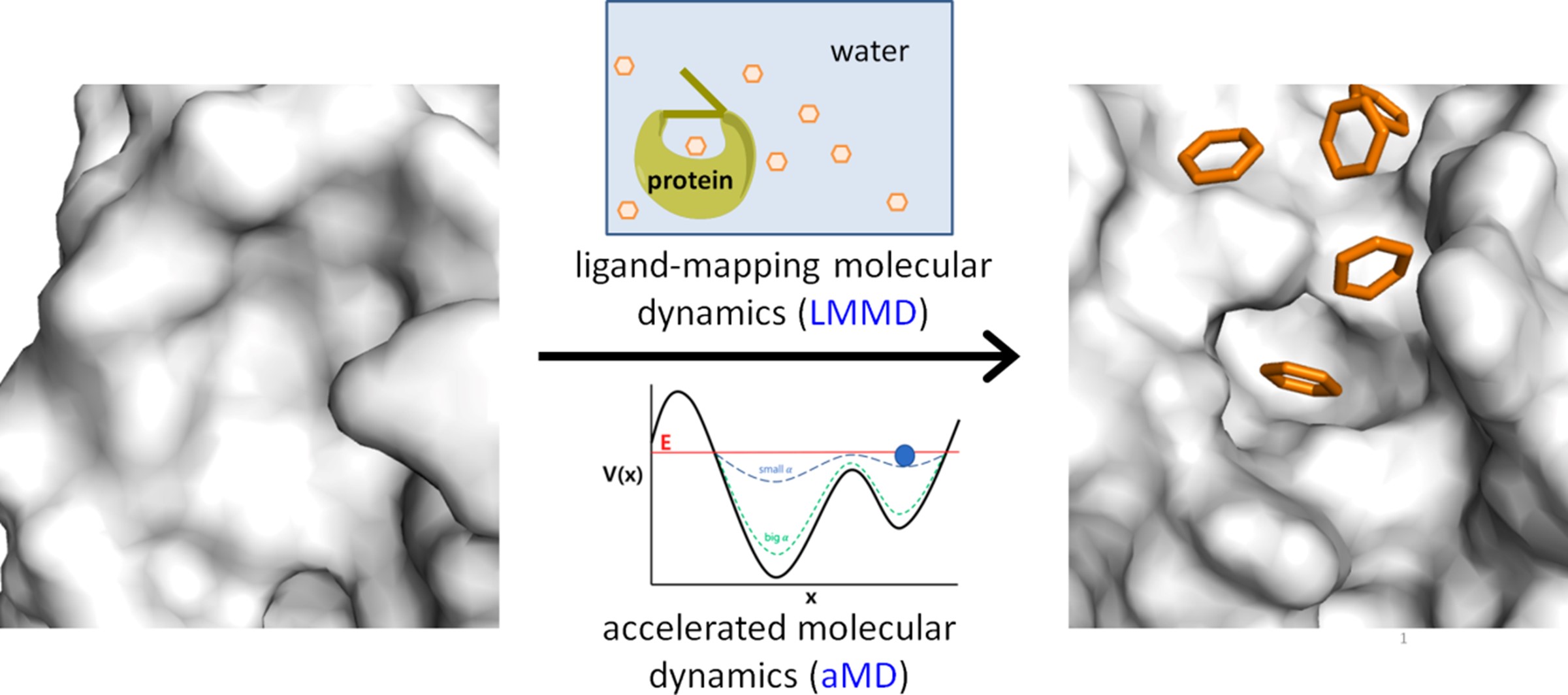
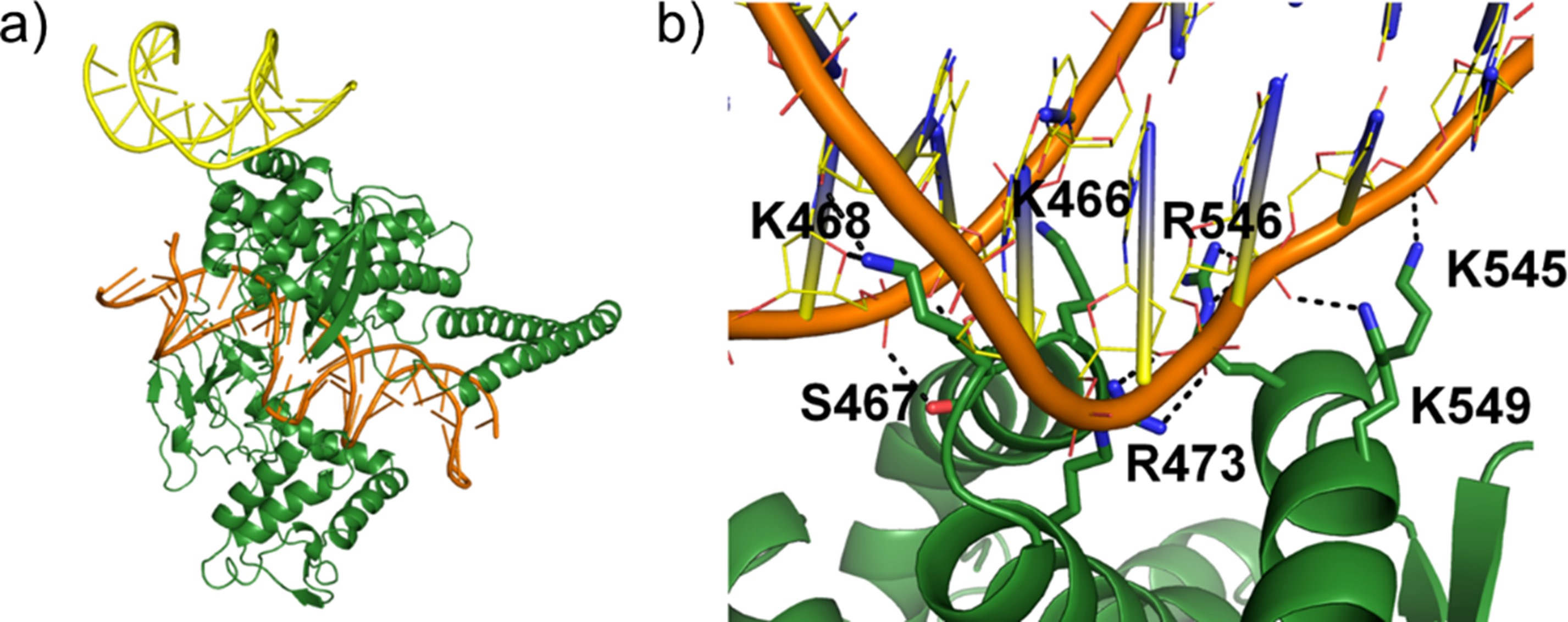
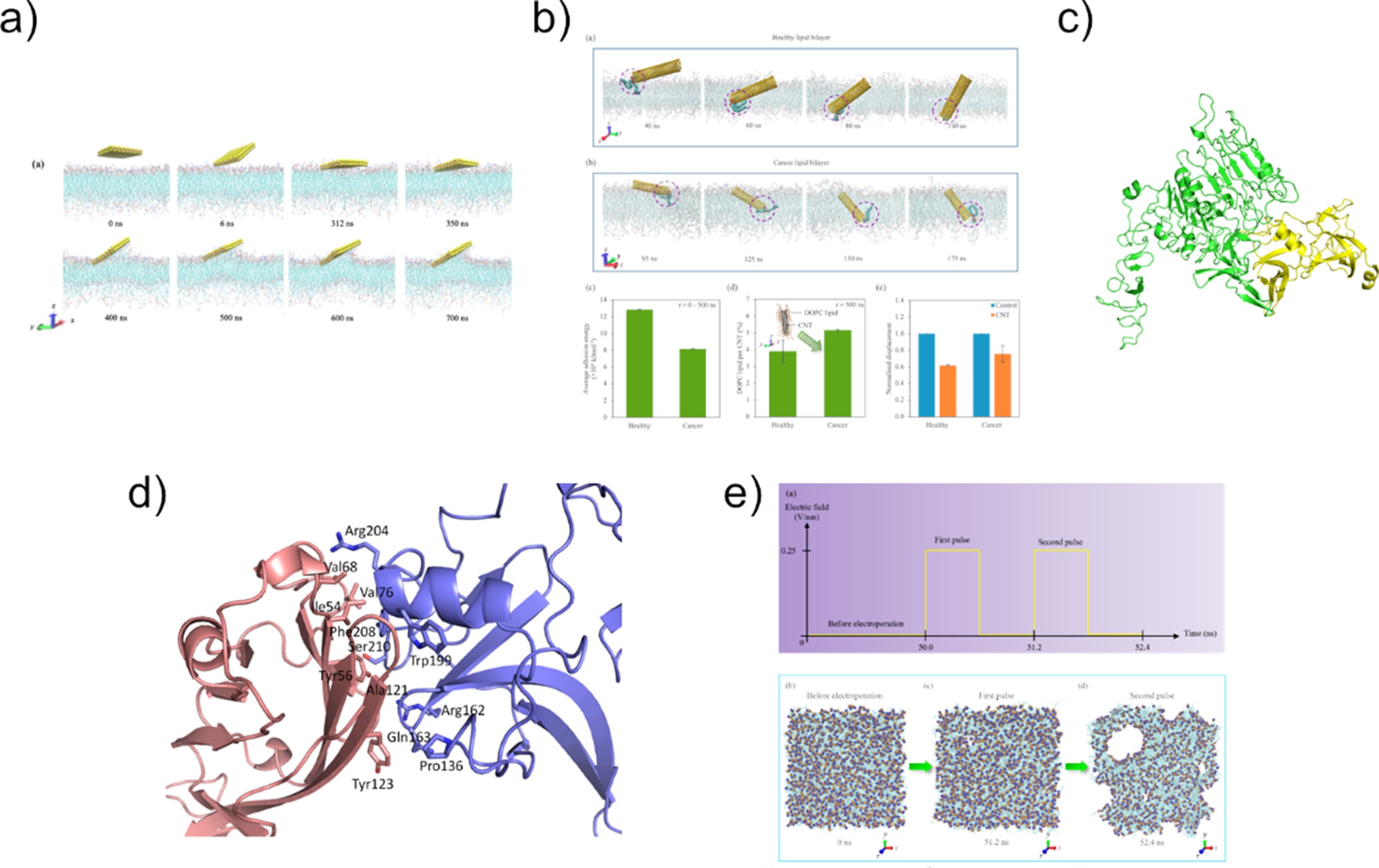
Dr. Tan’s research interests span the diverse fields of computational chemical biology, computer-aided drug design, and computational structural biology. He hopes to transform current strategies for drug discovery and chemical biology by improving the accuracy of binding site prediction, providing new insights into the structure and dynamics of drug targets, and guiding the rational design of therapeutics and chemical tools.
Group Members
| Scientist | NG Tze Yang Justin |
| Scientist | MODEE Rohit Laxman |
| Research Officer (T-UP with QDX) | CHONG Kian Chee |
| Research Officer | TAN Zi Ting Eunice |
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM