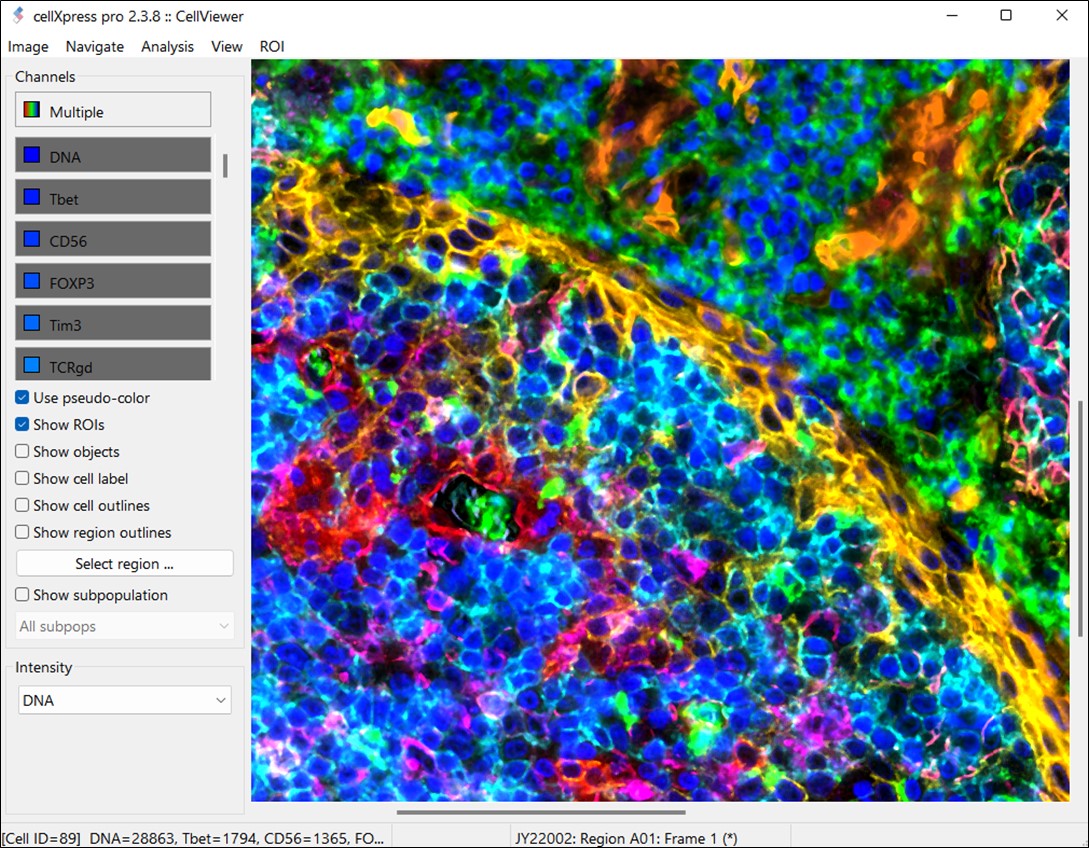
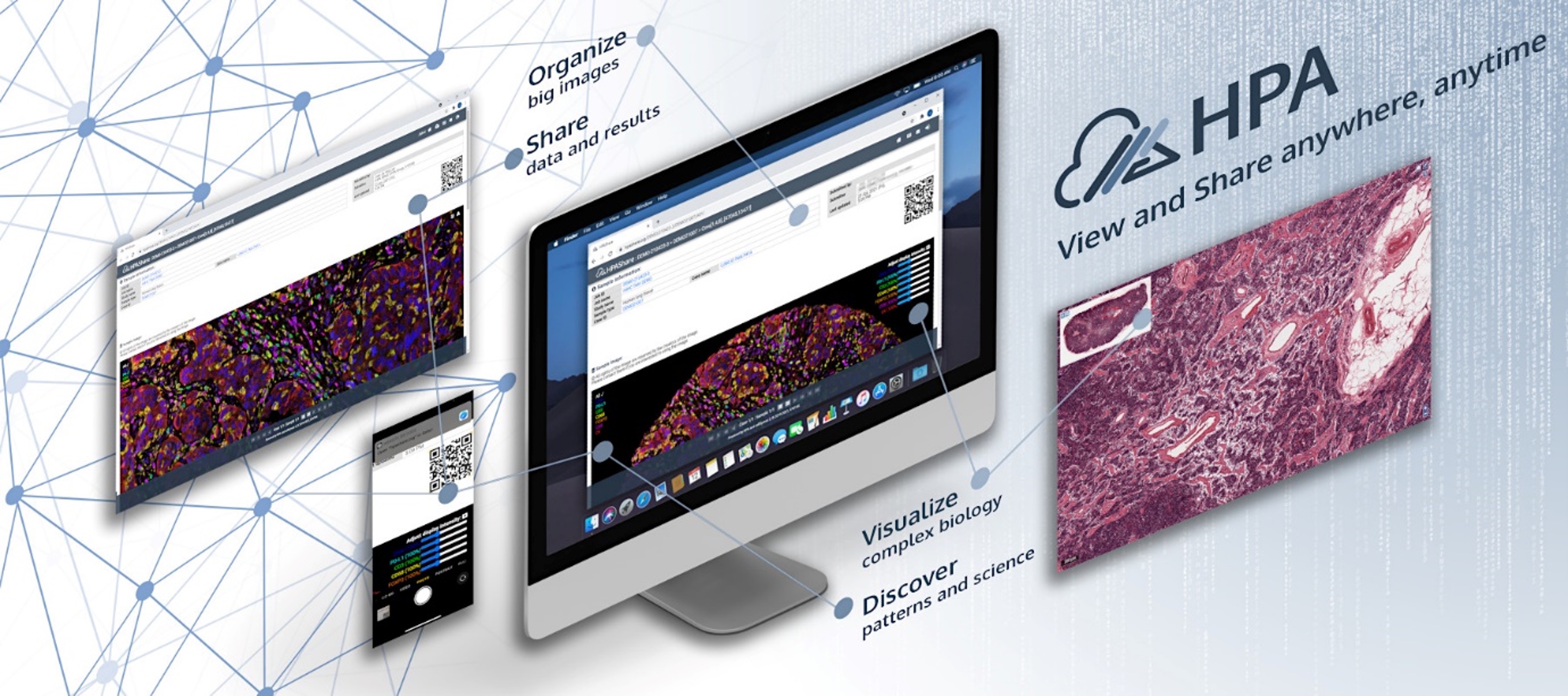
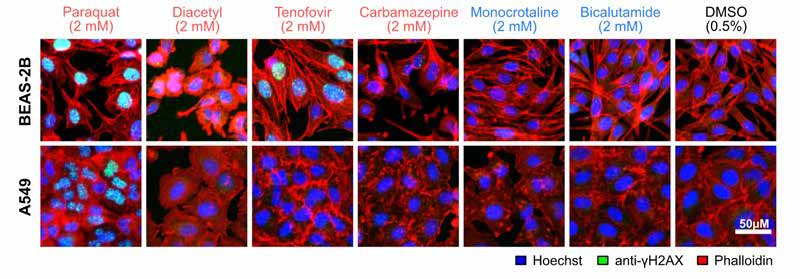
Complex Cellular Phenotype Analysis

Loo Lit Hsin
 | LOO Lit Hsin Senior Principal Investigator Email: loolh@a-star.edu.sg Research Group: Complex Cellular Phenotype Analysis |
Dr. Loo Lit Hsin is a Senior Principal Investigator at the Bioinformatics Institute (BII), A*STAR, Singapore. He is also an adjunct Assistant Professor at the Department of Pharmacology, Yong Loo Lin School of Medicine, National University of Singapore. Dr. Loo’s background is in computational and systems pharmacology/toxicology. He was the recipient of the Lush Prize – Science Award (2016), Award for Excellence in Postdoctoral Research (2010) and the Alfred Gilman Award (2009) by the University of Texas Southwestern (UTSW) Medical Center, and the Doctoral Award in Mathematical Sciences and Engineering (2005) by Drexel University. Dr. Loo was a postdoctoral fellow in the Bauer Center for Genomics Research at Harvard University (2005), and then in the Department of Pharmacology at the UTSW Medical Center, USA (2005-2010).
Group Members
| Lead Research Officer | LEE Jia Ying Joey |
| Senior Research Officer | KONG Jia Wen Carmen |
| Research Officer | GONZALES Edward Mark |
| PhD Student | YEO Chyi Maey Claresta |
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM