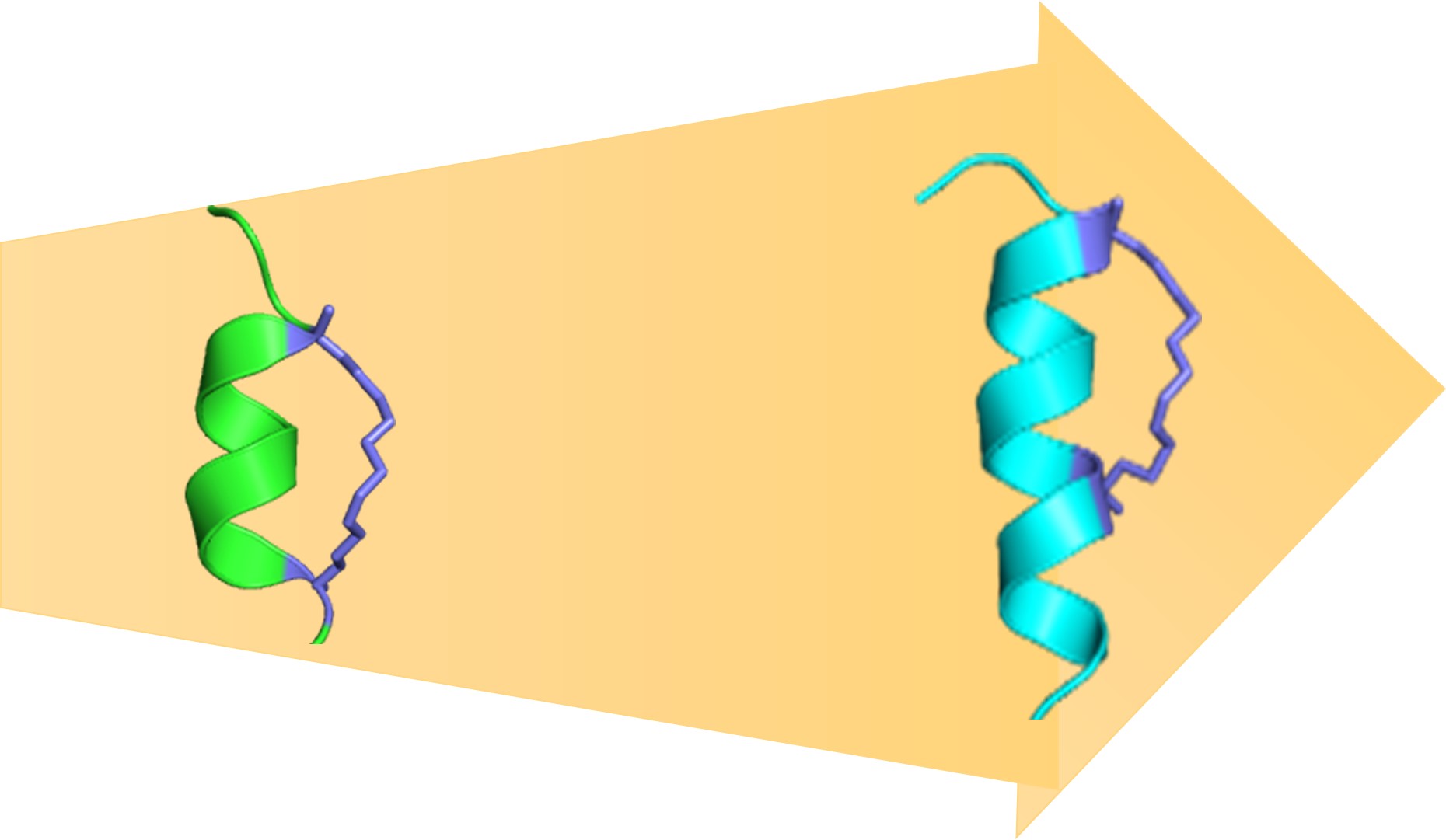
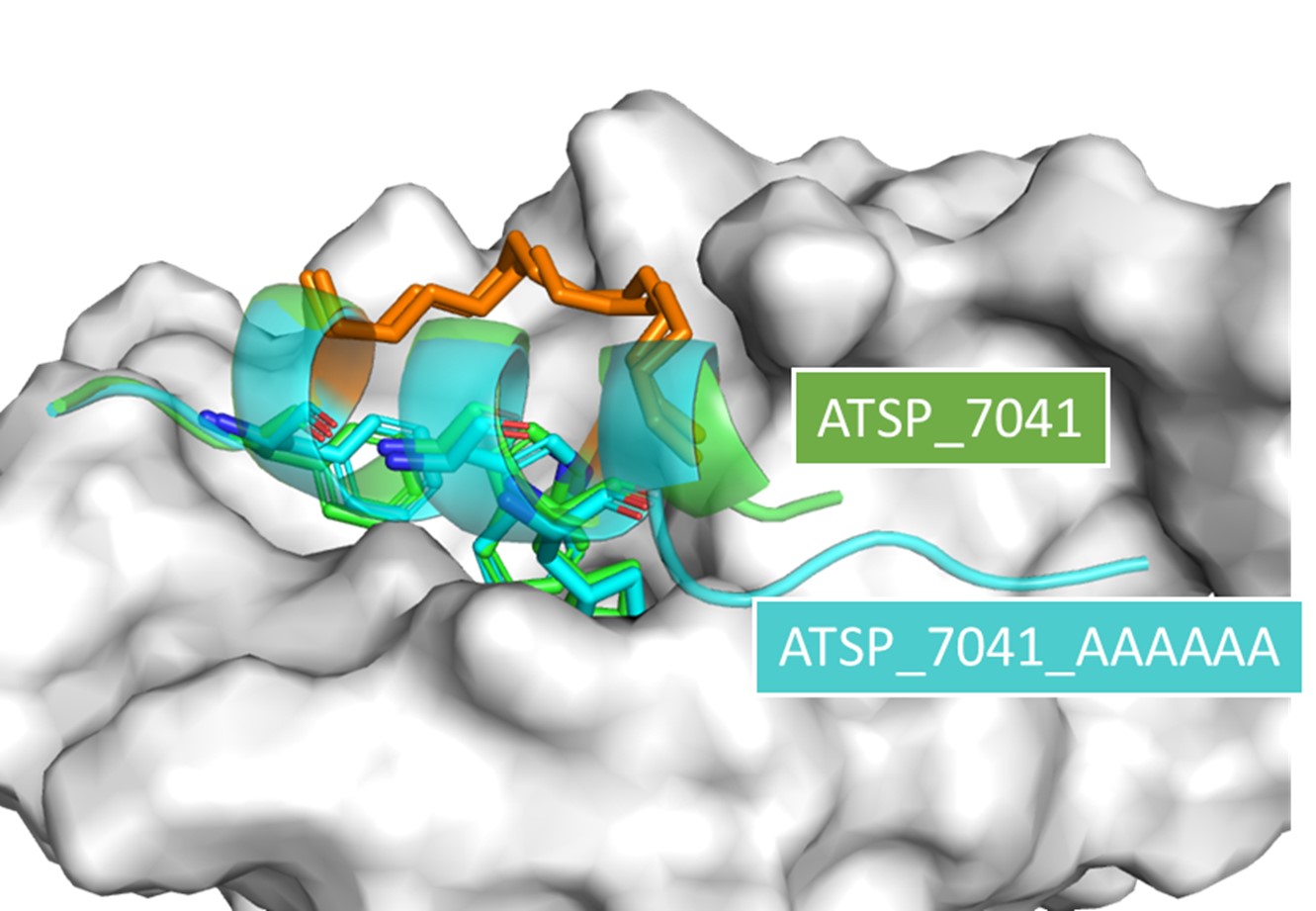
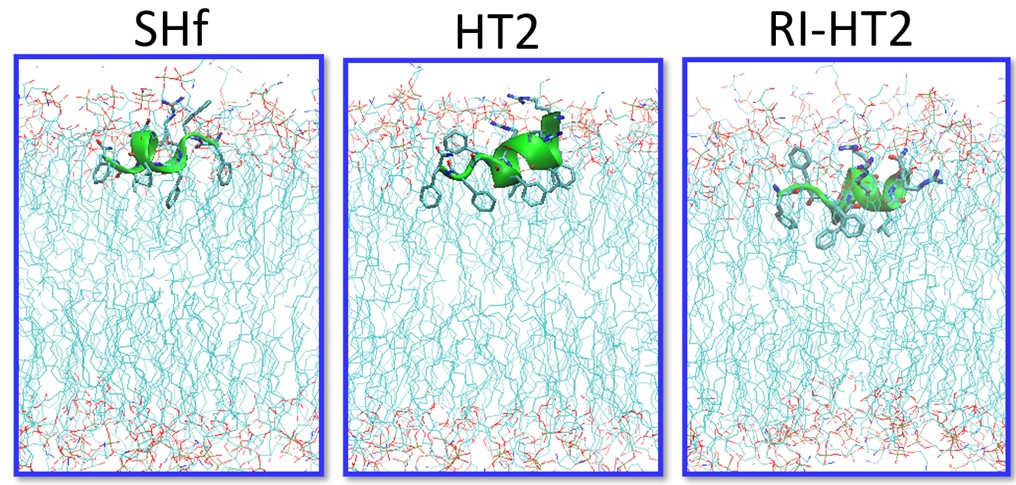
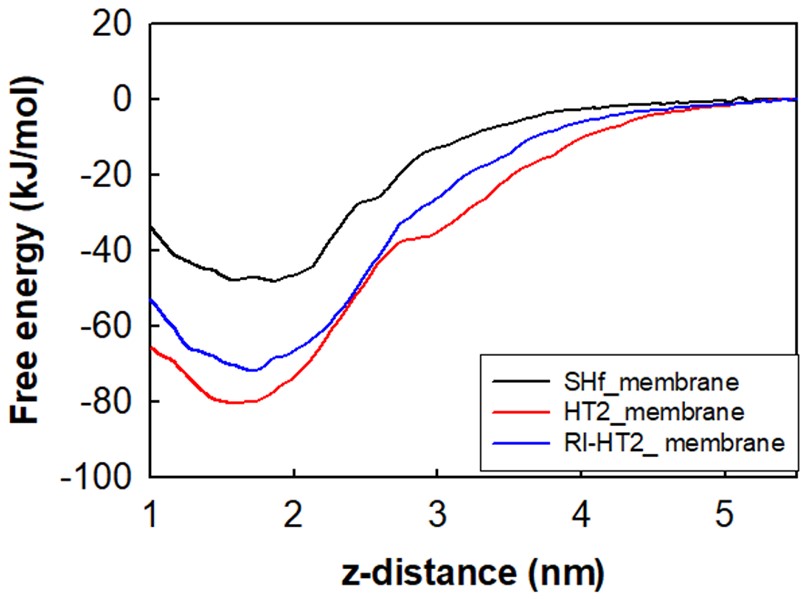
Atomistic Simulations and Design in Biology

Verma Chandra S.
 | VERMA Chandra S. (Head) Deputy Director (Research), Senior Principal Scientist III Email: chandra@a-star.edu.sg Research Group: Atomistic Simulations and Design in Biology |
Dr. Chandra Verma joined the Bioinformatics Institute (BII) A*STAR, Singapore, in November 2003. He heads the division of Biomolecular Modelling and Design and leads a group that applies physics-based models to understand the links between protein sequence, structure and biological function. His group works closely with experimental laboratories where the hypotheses generated are tested. In addition, the group is also involved in designing peptides and small molecules (through virtual screening) both for interrogating biology as well as for therapeutic purposes. Prior to joining Singapore, he worked at the Structural Biology Laboratory in York, UK. He obtained his undergraduate degree at the Indian Institute of Technology, India and his D. Phil at the University of York. He has cofounded two spinoffs: 1) Sinopsee Therapeutics - Start-up company that combines a yeast-based platform and modelling with colleagues from A*STAR IMCB and BTI to screen for and develop new molecules against targets in oncology and opthalmology. Series A funding is being explored for a novel potential therapeutic (eye drop) and 2) Aplomex - Start-up company that design small molecules, peptides, proteins/enzymes and antibodies.
Group Members
| Principal Scientist | SRINIVASARAGHAVAN Kannan |
| Principal Scientist | LI Jianguo |
| Principal Scientist | NGUYEN Ngoc Minh |
| Senior Scientist (T-up with Aplomex) | ARONICA Pietro |
| Senior Scientist | CHEN Tianqi |
| Scientist | SHIVGAN Aishwary |
| Scientist | LEE Chu Yin Bernadette |
| Research Officer | JAMAL Shamieraah |
| Research Officer | HIGHFIELD Christian |
| Research Officer | NGUYEN Hung Pham |
| Research Officer | SAMUEL Angel |
| PhD Student | YEO Hui Qing |
| PhD Student | LAM Joshua |
| PhD Student | SYME Tom |
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM