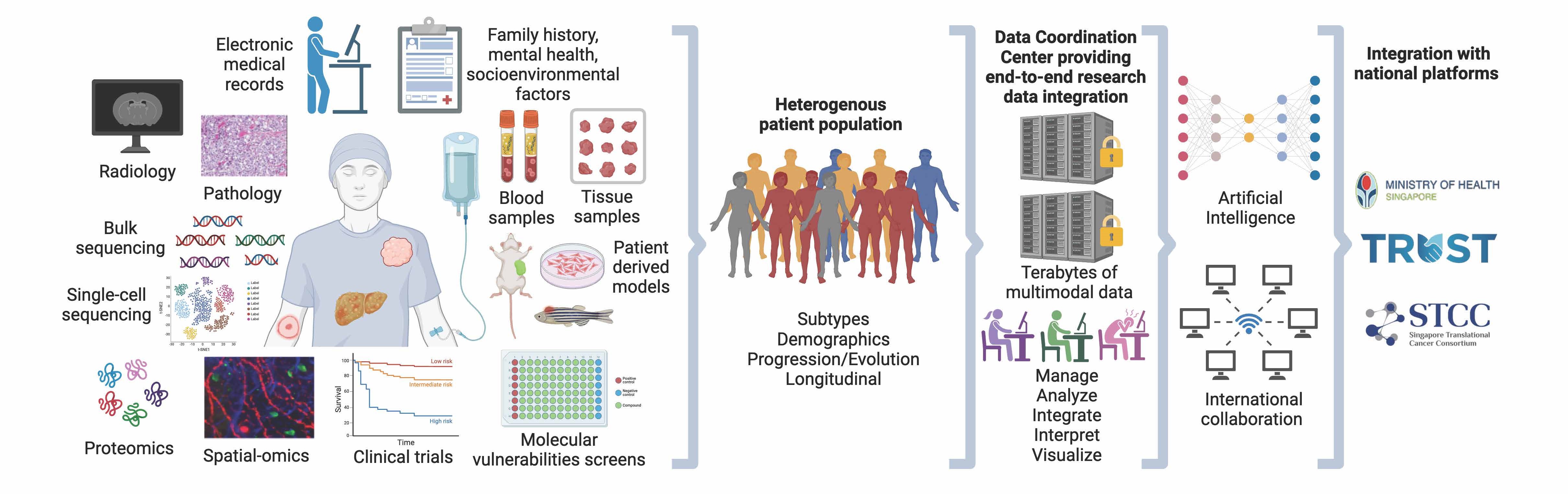
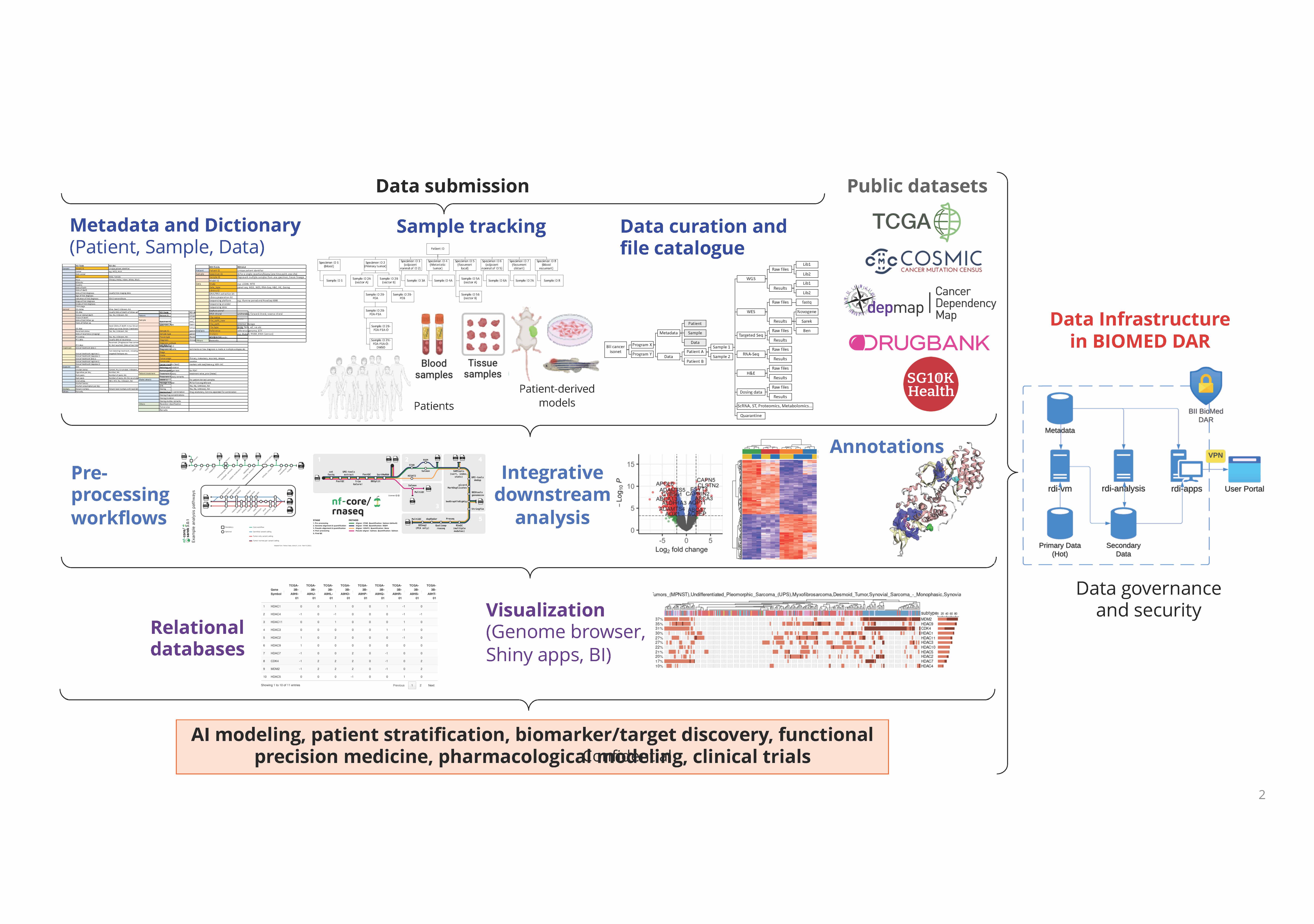
RESEARCH DATA INTEGRATION

Woo Xing Yi
 | WOO Xing Yi Head of Research Data Integration Email: woo_xing_yi@a-star.edu.sg Research Group: Research Data Integration |
Dr. Woo Xing Yi obtained her PhD in Chemical Engineering from the Joint-PhD program of University of Illinois at Urbana Champaign and National University of Singapore. She did her postdoctoral fellowship in the Genome Institute of Singapore under the mentorship of Professors Edison Liu and Guillaume Bourque. Subsequently, she worked at The Jackson Laboratory as a computational scientist and participated in a wide range of computational work, including analysis pipeline development, clinical curation and downstream analysis of large-scale datasets of cancer and patient-derived xenografts. She has led analysis projects in cancer resources and consortiums, and contributed to significant publications in high impact journals, including Nature Genetics, Proceedings of the National Academy of Sciences, Cancer Research and Genome Research.
Group Member
| Scientist | RUANO San Martin Camino |
| Scientist | NG Win Pin |
| Scientist | TAN Qiao Wen |
| Scientist | UMA Sangumathi Kamaraj |
| Research Officer | PHUA Xuan Ming Brandon |
| Research Officer | CHEN Junqi |
| Research Officer | TONG Haopeng |
| Research Officer | CASTILLO Francis Hans Napay |
A*STAR celebrates International Women's Day

From groundbreaking discoveries to cutting-edge research, our researchers are empowering the next generation of female science, technology, engineering and mathematics (STEM) leaders.
Get inspired by our #WomeninSTEM